1. Background
Hepatitis C virus (HCV) is a blood-borne pathogen that has infected about 170 million people worldwide (1). There are several modes of HCV transmission including blood transfusion, intravenous drug abuse, tattooing, transplantation from an infected donor, high-risk sexual exposure and birth to an infected mother. Using surgical and dental equipment which are not appropriately disinfected and sterilized can be additional sources of HCV transmission (2).
Six major HCV genotypes and more than 90 subtypes have been indentified thus far (3). HCV genotype has clinical importance as it determines the treatment duration with the current standard anti-HCV agents, pegylated interferon-α and ribavirin. Infections with genotype 1 are more challenging to treat with this medication regimen (4, 5). Distribution of HCV genotypes has also epidemiological significance. Genotypes 1, 2 and 3 are responsible for HCV infections in more than 90% of cases in North and South America, Europe and Japan (6). Genotypes 4 and 5a are frequent genotypes in Central Africa and South Africa respectively, whereas genotypes 1 and 2 are responsible for most of the HCV infections in West Africa. In Asia, the most frequent genotypes are 3 and 6 (7). HCV genotype 6 is generally restricted to Southeast Asia (8). In the Middle East, whereas, 4a is the predominant genotype in Arab countries, 1b and 3 are the most frequent genotypes in Turkey and Pakistan respectively (9, 10). The predominant genotype in Iran is 1a followed by 3a and 1b (11, 12, 13, 14). This paper reports the first ever case of HCV subtype-6a infection in Iran as no genotype-6 infection has been reported previously.
2. Case Presentation
A 62-year-old man with positive anti-HCV antibody was referred to Taleghani Hospital (Tehran, Iran). The patient has had no history of intravenous drug abuse, high-risk sexual activity nor has he shared shaving razors. The patient had a history of periodontal procedures, lower and upper gastrointestinal endoscopy. He underwent spine surgery in Hong Kong in 1977. The patient has had a Helicobacter pylori infection for 2 years. He has also been suffering from Parkinson’s disease for the last ten years. He has made trips to Britain (in 1991) and the United States (in 2010). He has had a history of admission to the hospital for respiratory difficulties. Serological tests for hepatitis B surface antigen (HBs-Ag) and antibodies to hepatitis A (IgM), hepatitis B core antigen (anti-HBc Ab) and hepatitis C virus (anti-HCV Ab) were performed using enzyme-linked immunosorbent assay (DIAPRO, Milan, Italy). Laboratory test results are shown in Table 1 .
aAbbreviations: ALT, Alanine aminotransferase; AST, aspartate aminotransferase; ALP, Alkaline phosphatase; N, Negative; P, Positive.
Viral RNA was extracted from patient’s plasma using QIAamp Viral RNA mini kit (QIAGEN, Hilden, Germany). HCV viral load was determined using HCV Real Time RT-PCR kit (Liferiver, Shanghai, China). Complementary DNA was synthesized using random hexamer primers and RevertAid Reverse Transcriptase (Fermentas, Vilnius, Lithuania) according to the manufacturer’s protocol. The 5’-UTR/Core region was amplified using PCR with primers NCRF: 5’- GGAACTACTGTCTTCACGCAGAAAGC -3’ and NCRR: 5’- GAAGCCGCACGTAAGGGTATCG -3’. Reaction conditions were 5 min at 95°C, followed by 35 cycles of 60s at 94°C, 40s at 63 °C and 60s at 72°C, with a final extension step for 7 minutes at 72°C. PCR product was direct sequenced using amplification primers. Phylogenetic tree was constructed using Molecular Evolutionary Genetic Analysis (MEGA) version 4.
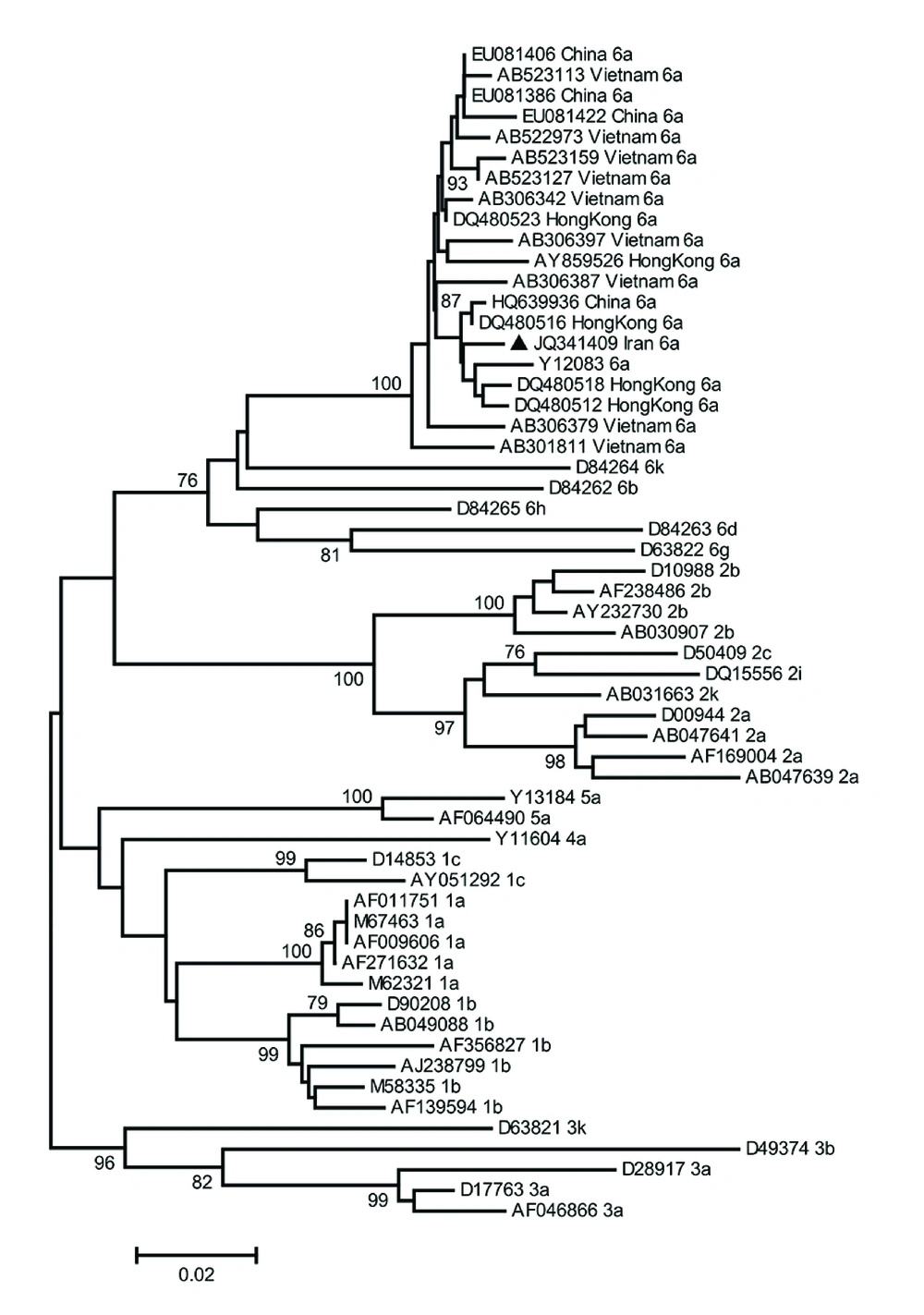
HCV RNA level was 9,572,718 IU/mL. The 5’-UTR/core sequence of the HCV isolate was compared with the corresponding region of the reference sequences retrieved from GenBank database. Genetic distances were calculated using Kimura two-parameter algorithm (15). The mean inter-genotypic distance between the 5’-UTR/core sequence of the HCV isolate obtained from the patient (GenBank accession number JQ341409) and corresponding regions of other genotype 6 isolates was 3.7%. The mean percentage of the inter-genotypic distance of the mentioned sequence from other genotypes was estimated to be 12.4-15.7 (Table 2). Phylogenetic tree was constructed and assessed by a bootstrap resampling test with 1000 replicates using MEGA version 4 (Figure 1). In the phylogenetic tree, the subject nucleotide sequence is classified as HCV subtype 6a.
| Genotype | IR a | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|---|
| 1 | 12.48 | |||||
| 2 | 15.32 | 15.15 | ||||
| 3 | 15.62 | 14.15 | 18.64 | |||
| 4 | 13.90 | 10.61 | 14.76 | 15.70 | ||
| 5 | 15.70 | 10.79 | 15.46 | 13.70 | 12.09 | |
| 6 | 3.70 | 12.59 | 15.55 | 15.61 | 14.70 | 14.96 |
aIranian isolate
3. Discussion
Distribution of HCV genotype 6 is mainly restricted to Southeast Asia and it is a rare or low-prevalence type in other parts of the world (16). Studies indicate that HCV genotype 6 is responsible for 50% of HCV infections in Myanmar and Vietnam (17, 18). Moreover, genotype 6 is the second most common HCV genotype among blood donors in Hong Kong (19). It should be mentioned that in the United States, one-third of HCV-positive immigrants from Southeast Asia are infected with various subtypes of genotype 6. Furthermore, HCV-6 has been observed in Cambodian and Vietnamese immigrants in Canada (16).
Previous studies have shown that the predominant HCV subtype in Iran is 1a followed by 3a. Other genotypes that have been identified in Iran are 1b, 2 and 4 (11-14). Samimi-rad et al. in 2004 studied 125 Iranian patients. They reported the existence of genotypes 1a, 3a, 1b and 4 in the studied population (11). Kabir et al. in 2006 reported subtypes 1a, 3a and 1b from 156 studied patients (12). In a different study in 2007, 2231 hepatitis C patients were tested by Keyvani et al. and subtypes 1a, 3a and 1b were detected (13). In 2010, Ziyaeyan et al. isolated HCV genotypes 3a, 1a, 1b and 2 from 634 studied individuals (14).
In this paper, we are reporting the first HCV genotype-6 infection diagnosed in an Iranian patient. The patient has had a history of multiple risk factors including a spine surgery in Hong Kong in 1977. Genotype 6 is currently the second most common genotype in Hong Kong. Due to the fact that HCV was identified in 1989 and the blood-screening programs started in the 1990’s, there is limited data on HCV epidemiology and geographical distribution of its genotypes prior to this time. Tanaka et al. in 2006, used the coalescent theory to investigate the past population dynamic of some HCV subtypes sampled from various countries including subtype 6a from Hong Kong. They showed that the exponential growth of HCV-6a population in Hong Kong occurred in the late 1970’s (20). Although the patient travelled to the United States in 2010, he has not articulated any high-risk exposure to any infectious sources of HCV during his trip. The data suggests that the spine surgery in Hong Kong in 1977 may have been the source of the HCV transmission in this particular case.
The current standard therapy for hepatitis C infection is pegylated interferon alpha in combination with ribavirin. The treatment duration and outcome typically depends on the HCV genotype. Whereas HCV genotypes 2 and 3 are responsive to the therapy in 70-80% of cases, genotypes 1 and 4 are considered to be difficult to eradicate with sustained virological response (SVR) rates around 50% (4, 5, 21). The SVR rate for genotype-6 infection is higher than that seen in genotype 1 infection but lower than that of genotypes 2 and 3 (22-24). Due to these facts, determining HCV genotype prior to considering a specific therapy protocol is of utmost importance. Iran is a low prevalence country in terms of HCV infection.
Due to increasing globalization, it is possible to encounter HCV genotypes or subtypes not previously reported in a specific geographical region. In this regard, diagnostic centers must consider the possibility of infections with genotypes that are rare or not previously reported in their region. Epidemiological studies are important to investigate different HCV genotypes distribution in various countries. However, completely relying on epidemiological data may prevent clinical diagnostic laboratories from considering HCV genotypes other than the common categories. In some cases, the patient may have been infected by a HCV genotype that was not observed in a region/country. If a commercial diagnostic kit or an in-house technique for HCV detection cannot detect such genotypes, it would produce a false negative result. The patient would thereby be a HCV carrier without a proper diagnosis and consequently can cause the spread of HCV in their family and eventually in their community. This being a case in point for the discussed issue highlights the fact that Iranian laboratories and physicians must not rule out the possibility of the HCV genotype-6 infection. While in more than 90% of hepatitis C cases in Iran, subtypes 1a, 3a or 1b are responsible for the infection, laboratories must consider HCV diagnostic techniques and materials (i.e. primers and molecular probes) by which all HCV genotypes can be detected.
In view of this report, further screening of groups with high-risk behavioral backgrounds, especially people who have travelled to foreign countries (particularly endemic regions) is needed. It is advised there be more a precise determination of the prevalence of various HCV genotypes/subtypes in Iranian population.