1. Background
Resistance in enteric Gram-negative bacteria, including Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and etc, is a matter of great concern because of growing numbers of reports of the resistant Gram-negative bacteria to all available antibacterial agents used in therapy. Extended spectrum beta-lactamase (ESBL) producer pathogens are resistant to penicillin, cephalosporin and monobactam but are not to carbapenem. These enzymes are made by Enterobacteriaceae, which aren’t active against cephamycins and imipenem, and demonstrate co-resistance to many of antibiotics (1).
The increase in ESBL-producer enteric Gram-negative bacteria has led to the choice of inappropriate therapy; as a result, the rate of resistance has increased (2). Antibiotic therapy of infections due to ESBL-producing pathogens still is a clinical challenge (3). For most of them, carbapenems and fluoroquinolones have been used (4). Among Gram-negative bacteria, E. coli is the most common pathogen in hospitals followed by K. pneumonia, Entrobacter, P. aeroginosa, Acinetobacter, Proteus mirabilis and Citrobacter (5).
P. aeruginosa was usually found in the medical solution, the aerators and traps of sinks, respiratory therapy equipments, bronchoscopes (6), on showerheads, and the surface of many types of raw fruits and vegetables (7). Taking imipenem has been identified as a risk factor for imipenem-resistant P. aeruginosa in some studies (8-10). A. baumannii can be found in both moist and dry environments, for instance, ventilator tubes, suction catheters, humidifiers, containers of distilled water and urine (11). There has been a considerable problem with ESBL producer Gram-negative bacteria, so this research was conducted to determine prevalence and antimicrobial susceptibility pattern of these organisms at a hospital in Tehran, Iran.
2. Objectives
The aim of this study was to determine prevalence and antimicrobial susceptibility pattern of ESBL and non-ESBL producing enteric Gram-negative bacteria.
3. Materials and Methods
A descriptive study was conducted in a private hospital in Tehran, Iran, between October and December in 2011. Patients’ samples were been obtained and were inoculated on blood agar (Oxoid Ltd, Bashingstone, Hampire, UK) and MacConkey agar (Oxoid Ltd, Bashingstone, Hampire, UK plates at microbiology laboratory hospital. We incubated these plates at 37°C for 24 hours, and recognized significant isolates as species level by common bacteriological techniques. Totally, 292 Gram-negative species were isolated at Microbiology Laboratory from patients’ specimens. Control organisms E. coli ATCC 35218; K. pneumoniae ATCC 700603; P. mirabilis ATCC 35659; P. aeruginosa ATCC 27853; Serratia marcescense ATCC 43862; and A. baumannii ATCC BAA-747 were used to control the quality of media and evaluate color stability. Three control American Type Culture Collection (ATCC) organisms were used for each test, and all of them were tested for each new group varied.
Susceptibility pattern was determined by disk diffusion method based on Kirby-Bauer method on Mueller-Hinton agar (Oxoid Ltd, Bashingstone, Hampire, UK) plate (12). The following disks were been used: Amoxicillin-clavulanate (30 μg/disk) was been placed in the center of plate while ceftriaxone (30 μg/disk), cefixime (5 μg/disk), cefotaxime (30 μg/disk), carbenicillin (100 μg/disk), cephalexin (30 μg/disk), trimethoprim-sulfamethoxazole (25 μg/disk), amikacin (30 μg/disk), gentamicin (10 μg/disk), ciprofloxacin (5 μg/disk), and imipenem (10 μg/disk), tetracycline (30 μg/disk), nalidixic acid (30 μg/disk), piperacillin-tazobactam (110 μg/disk), and colistin (10 μg/disk) were been located around of the central disc (Oxoid Ltd, Bashingstone, Hampire, UK). Five disks were been placed on each plate. The CLSI recommends a zone size of ≥ 5 mm difference between the cephalosporin disk with and without clavulanic acid is considered as significant and ESBL producing (13). SPSS 16 software (descriptive analysis, Chi-square) was used for statistical analysis of this study. A P value of < 0.05 was considered to be statistically significant.
4. Results
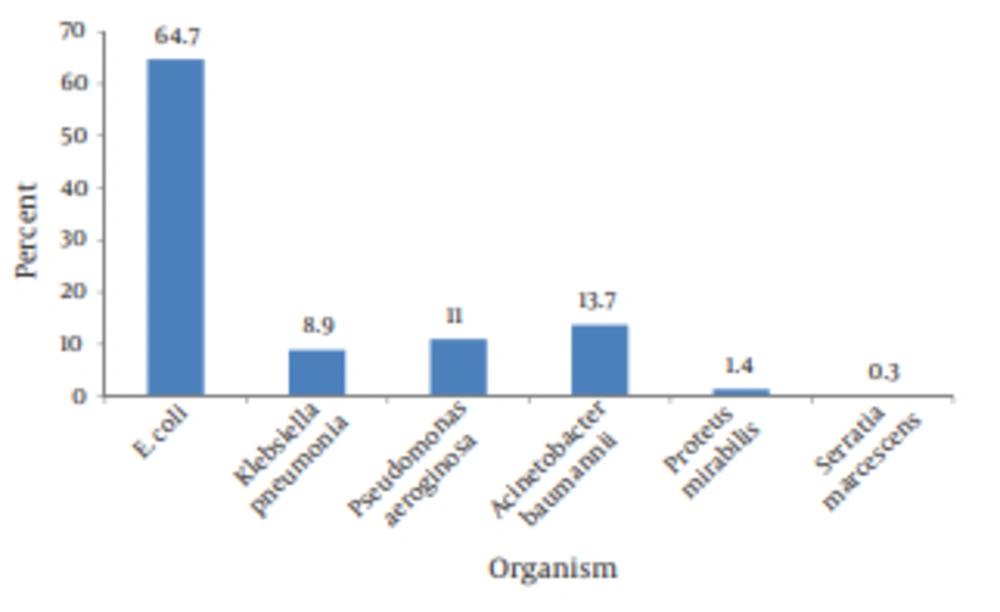
155 of patients were female (53.1%) and 137 were male (46.9%). The mean age of patients was 65.42 ± 19 years .The age of the oldest patient was 98 and youngest one was 1. The majority of patients younger than 35 years and older than 55 years old were females. In terms of the source of isolates, urine accounted for the greatest number [207 (71%)], followed by sputum and tracheal secretion [62 (23.9%)], wound [12 (4.1%)], blood [7 (2.4%)], pleural fluid [2 (0.7%)], synovial fluid [one (0.3%)], and peritoneal catheter [one (0.3%)]. As shown in the Figure 1, E. coli was the most common organism [189 (64.7%)], Followed by A . baumannii [40 (13.7%)], P . aeruginosa [32 (11%)], K . pneumonia [26(8.9%)], P . mirabilis [4 (1.4%)], and S . marcescense [1 (0.3%)]. 122 (41.8%) out of 292 isolated Gram negative were ESBL–producing. Most of the ESBL-producing isolated were isolated from urine samples (N = 101), followed by sputum and tracheal secretion (n = 8), blood (n = 6), wound (n = 6), and synovial fluid (n = 1).
Resistant pattern of ESBL-producing and nonproducing isolated has been shown in Table 1. All of the ESBL producer isolates were sensitive to imipenem, piperacillin- tazobactam and colistin whilst they were resistance to cephalexin (96.7%), ceftriaxone (95.9%), cefotaxime (95.9%), cefixime (95.9%), carbenicillin (95.9%), ciprofloxacin (85.2%), trimethoprim-sulfamethoxazole (84.4%), tetracycline (78.6%), gentamicin (62.2%), nitrofurantoin (21.3%) and amikacin (10.6%). All isolates were sensitive to imipenem, colistin and piperacillin-tazobactam while 87 (71.3%) and 99 (81.1%) of isolates were sensitive to nitrofurantoin and amikacin, respectively.
In the non-ESBL producer group, 56 isolates (33%) were resistant to imipenem, 42 (25%) to piperacillin-tazobactam, and 2 (1%) to colistin. 14 (25%) and 42 (75%) out of 56 isolated resistant to imipenem were sensitive and resistant to piperacillin-tazobactam, respectively. Among 114 isolates resistant to nitrofurantoin, 26 (22.8%) and 88 (77.2%) were belonged to ESBL and non-ESBL groups, respectively.
| Antibiotic | ESBL-Producing | ||
|---|---|---|---|
| Yes | No | ||
| Nitrofurantoin | Sensitive | 87 | 74 |
| Intermediate | 9 | 8 | |
| Resistant | 26 | 88 | |
| Trimethoprim-Sulfamethoxazole | Sensitive | 19 | 25 |
| Intermediate | 0 | 0 | |
| Resistant | 103 | 118 | |
| Gentamicin | Sensitive | 43 | 109 |
| Intermediate | 3 | 3 | |
| Resistant | 76 | 58 | |
| Tetracycline | Sensitive | 26 | 37 |
| Intermediate | 0 | 1 | |
| Resistant | 96 | 132 | |
| Piperacillin-Tazobactam | Sensitive | 122 | 126 |
| Intermediate | 0 | 2 | |
| Resistant | 0 | 42 | |
| Colistin | Sensitive | 122 | 168 |
| Intermediate | 0 | 0 | |
| Resistant | 0 | 2 | |
| Ciprofloxacin | Sensitive | 18 | 103 |
| Intermediate | 0 | 1 | |
| Resistant | 104 | 66 | |
| Imipenem | Sensitive | 122 | 112 |
| Intermediate | 0 | 2 | |
| Resistant | 0 | 56 | |
| Amikacin | Sensitive | 99 | 120 |
| Intermediate | 10 | 5 | |
| Resistant | 13 | 45 | |
| Ceftriaxone | Sensitive | 5 | 85 |
| Intermediate | 0 | 3 | |
| Resistant | 117 | 82 | |
| Cefotaxime | Sensitive | 5 | 85 |
| Intermediate | 0 | 3 | |
| Resistant | 117 | 82 | |
| Cephalexin | Sensitive | 4 | 72 |
| Intermediate | 0 | 2 | |
| Resistant | 118 | 96 | |
| Cefixime | Sensitive | 5 | 80 |
| Intermediate | 0 | 4 | |
| Resistant | 117 | 86 | |
| Carbenicillin | Sensitive | 5 | 89 |
| Intermediate | 0 | 1 | |
| Resistant | 117 | 80 | |
| Total | 122 | 170 | |
Interpretive results of nitrofurantoin susceptibility testing of ESBL and non-ESBL producer isolates with phenotypes for resistance and isolates with phenotypes for susceptibility to trimethoprim-sulfamethoxazole, ciprofloxacin, the third generation cephalosprins (ceftriaxone, cefixime, and cefotaxime), gentamicin, amikacin and imipenem were shown in Tables 2 and 3. More than 70% of the ESBL isolates of resistant to trimethoprim-sulfamethoxazole, ciprofolaxacin, the third generation cephalosprins, gentamicin and amikacin, were sensitive to nitrofurantoin whereas most non-ESBL producer isolates, which were resistant to the above antibiotics, were also resistant to nitrofurantoin. Furthermore, 98.2% of imipenem-resistant non-ESBL isolates were resistant to nitrofurantoin. We found no statistically significance in our study.
| Isolates with indicated susceptibility to nitrofurantoin, % | |||
| Isolate phenotype | Sensitive | Intermediate | Resistant |
| Trimethoprim-Sulfamethoxazole | |||
| Sensitive | 74 | 0 | 26 |
| Intermediate | 0 | 0 | 0 |
| Resistant | 70.9 | 8.8 | 20.3 |
| Ciprofloxacin | |||
| Sensitive | 66.6 | 0 | 33.4 |
| Intermediate | 0 | 0 | 0 |
| Resistant | 72.1 | 8.5 | 19.4 |
| The third generation cephalosporin a | |||
| Sensitive | 80 | 0 | 20 |
| Intermediate | 0 | 0 | 0 |
| Resistant | 70.9 | 7.6 | 21.5 |
| Gentamicin | |||
| Sensitive | 69.7 | 4.7 | 25.6 |
| Intermediate | 100 | 0 | 0 |
| Resistant | 71 | 9.2 | 19.8 |
| Amikacin | |||
| Sensitive | 71.7 | 6.1 | 22.2 |
| Intermediate | 70 | 20 | 10 |
| Resistant | 69.3 | 7.6 | 23.1 |
| Imipenem | |||
| Sensitive | 71.3 | 7.4 | 21.3 |
| Intermediate | 0 | 0 | 0 |
| Resistant | 0 | 0 | 0 |
a Ceftriaxone, Cefixime, and Cefotaxime
| Isolates with indicated susceptibility to nitrofurantoin, % | |||
| Isolate phenotype | Sensitive | Intermediate | Resistant |
| Trimethoprim-Sulfamethoxazole | |||
| Sensitive | 80.8 | 11.5 | 7.7 |
| Intermediate | 0 | 0 | 0 |
| Resistant | 27.1 | 1.7 | 71.2 |
| Ciprofloxacin | |||
| Sensitive | 63.1 | 5.9 | 31 |
| Intermediate | 0 | 0 | 100 |
| Resistant | 13.7 | 3 | 83.3 |
| The third generation cephalosporin a | |||
| Sensitive | 77.7 | 7 | 15.3 |
| Intermediate | 0 | 33.3 | 66.7 |
| Resistant | 9.8 | 1.2 | 89 |
| Gentamicin | |||
| Sensitive | 61.5 | 6.4 | 32.1 |
| Intermediate | 66.7 | 0 | 33.3 |
| Resistant | 8.6 | 1.7 | 89.7 |
| Amikacin | |||
| Sensitive | 56.7 | 5.8 | 37.5 |
| Intermediate | 80 | 20 | 0 |
| Resistant | 4.4 | 0 | 95.6 |
| Imipenem | |||
| Sensitive | 65.2 | 6.3 | 28.5 |
| Intermediate | 0 | 50 | 50 |
| Resistant | 1.8 | 0 | 98.2 |
a Ceftriaxone, Cefixime, and Cefotaxime
5. Discussion
In this research, 122 (41.8%) of isolated Gram negative were ESBL-producing which most of them were E. coli [n = 112 (91.8%)], followed by K. pneumonia [n = 10 (8.2%)]. In Geng et al.’s study, the most common organisms were E. coli (47.4%) and K. pneumoniae (37.3%) (14). The similar data have been found in Alipourfard’s study (60% and 40%, respectively) (1) and Babypadmini’s research (41% and 40%, respectively) (15). In our study, all ESBL producer isolates were sensitive to imipenem, piperacillin-tazobactam and colistin, and the rates of susceptibility to these antibiotics in the non-ESBL group were 65.9%, 74.1% and 98.8%, respectively. Thus, the rates of imipenem, piperacillin-tazobactam and colistin resistant isolates in the non-ESBL producer group were higher than ESBL group.
In studies conducted by Geng, Akyar, Grandesso, and Kothari, the overall rates of susceptibility of ESBL producer E. coli to piperacillin-tazobactam were comparable to our research (14-17). The similar data about imipenem were found in Geng’s study (14). In Alipourfard et al’s study all ESBL producer Gram-negative bacteria were susceptible to carbapenems (1). Rafay et al. showed the lower sensitivity (49.6%) to piperacillin-tazobactam (18). In a study conducted by Olusolabomi et al, the ESBL producers were multiplying resistant and moderately sensitive to colistin (19).
In this research, resistance to the third generation cephalosprins (95.9%), ciprofloxacin (85.2%), trimethoprim-sulfamethoxazole (84.4%), gentamicin (62.2%), and amikacin (10.6%) were detected in the ESBL producer group. Susceptibility rate to amikacin was approximately six-fold higher than gentamicin. In the ESBL producer isolates, the rate of resistance to ciprofloxacin, gentamicin, trimethoprim-sulfamethoxazole and the third cephalosprins was very high (more than 70%). Alipourfard, Akyar, and Grandesso reported related findings about amikacin and ciprofloxacin (1, 15, 17). Rafay and Asha Pai have showed the similar data about ciprofloxacin and amikacin, respectively (18, 20).
Resistance to amikacin in the Geng’s study was higher (14). The similar data in resistant pattern to trimethoprim-sulfamethoxazole have been obtained in the researches conducted by Akyar, Grandesso, and Rafay (15, 17, 18). Alipourfard et al found the rate of resistance to the third generation cephalosprins was very high (1). Tekin and Karlowsky showed the more sensitivity to trimethoprim-sulfamethoxazole and ciprofloxacin than our study (21, 22). Based on the above findings, susceptibility rates of ESBL to the third generation cephalosprins, ciprofloxacin, gentamicin and trimethoprim-sulfamethoxazole have been decreased in the recent years.
In present study, rates of susceptibility to nitrofurantoin were 71.3% and 43.5% in the ESBL and non-ESBL producer isolates, respectively. Thus, the rate of resistance to nitrofurantoin in ESBL group was lower than of that in non- ESBL group. Alipourfard (57,4%), Kothari (65.7%), Rafay (57.4%) and Hamze (45%), reported the lower susceptibility to nitrofurantoin (1, 16, 18, 23), in contrast with Babypadmini (89%), Grandesso (89.3%), Asha Pai (88.94%), and Tekin (93.1%) (17, 20, 21, 24). Garau reported among isolates of ESBL producer E. coli, 71.3% (equal to our finding) were susceptible to nitrofurantoin. Garau recommended nitrofurantoin be considered an alternative, rather than a first-line, therapeutic agent for these organisms (25). Sometimes, we had limitations in preparing disks, cost, and instruments. In such studies, availability of expert laboratory employments is very important.
To sum up, although all ESBL producer Gram-negative bacteria are very sensitive to imipenem, piperacillin-tazobactam, and colistin, non-ESBL isolates showed resistant pattern. In present study, 70% of clinical isolates were obtained from urine and most ESBL isolates of this study were E. coli and the majority of the ESBL isolates (79%) were sensitive to nitrofurantoin. In addition, certain researchers recommend nitrofurantoin as an alternative therapeutic agent for lower urinary tract infections due to these organisms. Based on the above findings, it could be surmised that nitrofurantoin would be an appropriate antibiotic against lower urinary tract infection due to ESBL-E. coli.