1. Background
The microbial population in the dental plaque is very complex. Development of human periodontitis is particularly related to the number of normal oral flora (1). From more than 500 different types of oral cavity isolated bacteria (2), only a small proportion has been documented as periodontal pathogens (3, 4). The major pathogenic bacteria species in etiology of periodontal diseases are Aggregatibacter actinomycetemcomitans (formerly Actinobacillus actinomycetemcomitans) and Porphyromonas gingivalis (5, 6). Prevotella intermedia, Tannerella forsythensis and Treponema denticola are some of the oral normal flora associated to periodontal disease (7).
An inflammatory disorder, involving the supporting structures of the teeth is defined as periodontitis. Unfortunately, periodontitis damages are permanent and may expand to such a degree that complex and prolonged therapy is essential to control the condition. With decrease the incidence of dental caries in adults, periodontitis has become the most common cause for tooth loss (8). The bacterial species mentioned above are frequently isolated from the damaged periodontal sites (9-11), and it has been suggested that bacterial collaboration might promote the infectious process of periodontal disease. Consequently, it is important to know the composition of microorganisms in human periodontal pockets for diagnosis and rational treatment. Accordingly, it is important to recognize the composition of bacteria in human periodontal pockets for diagnosis and disease management (12).
In addition to culture based methods, techniques such as immunoassays, enzyme assays, and nucleic acid probe assays have been developed for the identification of some oral pathogens (13-17). Nevertheless, detection limit of the techniques is about between 103 to 105 targets per sample. Laboratory techniques have depth effect on detection of these putative pathogens in periodontal health and disease. Although classic method such as cultivation are used for oral microbial evaluations, but the microbial profile can be significantly underestimated because many bacteria (especially anaerobes) cannot be cultivated through these techniques. Also isolation and cultivation of the oral flora is accepted as the gold standard for many years (18).
Nowadays, polymerase chain reaction (PCR) is regarded as a vital tool for the rapid, sensitive, and specific detection of various bacterial and viral pathogens (19). Also real-time PCR has been proposed as a powerful diagnostic tool for rapid, sensitive and quantitative detection of the bacterial pathogens (20). Easy monitoring of periodontitis and more accurate epidemiological studies on the succession of periodontal disease are major consequents of developing an accurate non-culture based assay for detecting the oral pathogens (7).
2. Objectives
In this study, we designed and developed both PCR and SYBR Green real time PCR assays for immediate detection of A. actinomycetemcomitans and T. forsythensis then compared analytical sensitivity and specificity of the assays. Although several PCR-based methods for detection of the oral pathogens have been reported (18, 19, 21-26), we hereby report the first designed duplex PCR and real time PCR assays in Iran for sensitive, specific and rapid detection of the two pathogens. To address an improved PCR assay potentially capable for application in clinic, recommendations of CLSI (Clinical Laboratory Standards Institute) for evaluation and validation of the molecular assays such as analytic sensitivity and analytic specificity were noted.
3. Materials and Methods
3.1. The Bacterial Strains
The genome of wild type bacteria of A . actinomycetemcomitans strain HK1651 (ATCC 700685D-5) and T . forsythensis strain FDC 338 (ATCC 43037D-5) were used as positive control. Negative control bacteria were bought in lyophilized form explained in Table 1. The bacterial genomes were extracted using DNP™ kit (Cinnagen, Iran) then, the purity of culture was approved. The extracted genomes were quantified using a Picodrop Spectrophometer (Picodrop, UK) according to the manufacturer’s instructions.
| Organism Name | Strain Number | Supplier |
|---|---|---|
| Staphylococcus aureus | ATCC 25923 | |
| Bacillus subtilis | ATCC 6051 | |
| Shigella sonnei | ATCC 9290 | |
| Escherichia coli | ATCC 25922 | Pasture Institute of Iran |
| Enterococcus faecalis | ATCC 29212 | |
| Pseudomonas aeruginosa | ATCC 27853 | |
| Klebsiella pneumoniae | ATCC 7881 | |
| Yersinia enterocolitica | PTCC 1480 | Iranian Scientific and Industrial Research Organization |
| Yersinia pseudotuberculosis | PTCC 1244 | |
| Proteus vulgaris | PTCC 1079 | |
| Citrobacter freundii | PTC |
Specifications of the Negative Control Bacteria Used in the Study
3.2. Designing the Target Gene Primers
Hemoglobin-binding protein (hbpA) gene of A. actinomycetemcomitans and 16S rRNA gene of T. forsythensis were targeted for specific primer pair designing. All 83 NCBI deposited sequences of hbpA gene and 10 NCBI deposited sequences of 16S rRNA gene were found, downloaded and aligned by CLC Main Workbench software version 5.5 (The accession numbers not shown). Allele ID software version 7.60 was used for designing primers based on the two alignment resulted consensus sequence. The characteristics of the designed primer pairs are described in Table 2.
| Bacterium | Target Gene | Primer Name | Primer Sequence | Product Length, bp |
|---|---|---|---|---|
| Aggregatibacter actinomycetemcomitans | hbpA | F-Aggrega- hbpA | 5´- AGACCCAATGCAAAAGTAACG | 160 |
| R- Aggrega- hbpA | 5´- GCAGTTCTGGGCTGAATTG | |||
| Tannerella forsythensis | 16S rRNA | F- Tan-16 | 5´- GCATGTACCTTGTGAATAAGCA | 250 |
| R- Tan-16 | 5´- CTTCGCAATCGGAGTTCTG | |||
| 16S rRNA | 16S-350F, Universal primer | 5ʹ-CCTACGGGAGGCAGCAGT | 475 | |
| 16S-820R, Universal primers | 5ʹ-CGTTTACGGCGTGGACTAC |
Characteristics of the Designed Primer Pairs That Were Utilized for Molecular Detection of A. actinomycetemcomitans and T. forsythensis
3.3. Conventional PCR Experiment
Two PCR experiments were conducted in a volume of 25 µL, consisting 1 × PCR buffer (Fermentas, GmbH, Germany), 2 mM Mg 2+ , 0.2 mM dNTPs, 0.5 µM of F-Aggrega-hbpA, R- Aggrega-hbpA or F-Tan-16, R-Tan-16 primers pair (Table 2), 2 µL of A. actinomycetemcomitans or T. forsythensis genomic DNA (35 ng/µL) and 1 unit Taq DNA polymerase. Amplification using a certain thermal program was carried out for 35 cycles in the PCR program was set as follow: initial denaturation at 95°C for 5 minutes, denaturation at 95°C for 1minute, and annealing temperature at 60.8°C for 45 seconds and extension time at 72°C for 45 seconds. The annealing temperature optimization was done using gradient PCR ranged from 58 to 63°C. In addition a 25 µL reaction was amplified with the same condition but distilled water was used instead of bacterial genomes as the negative control. In order to determine the presence of amplicons, the PCR products were separated by electrophoresis on 2% agarose gel containing ethidium bromide intercalating dye. To improve the assays for simultaneous detection of the genes, a duplex PCR assay was set up. With a volume of 50 µL, the duplex PCR assay consisted of 3 mM Mg 2+ , 2 mM dNTPs, 0.2 µM of the both primers pairs and 1 unit of Taq DNA polymerase enzyme.
3.4. Development of Positive Control Sequences
The positive control is used to ensure that the PCR has been done and determine the band length of target sequence. Accordingly, cloning of PCR products of hbpA and 16S rRNA genes in plasmid vector was used in present study. TA cloning was done by InsTA clone PCR cloning kit (Fermentas, Lithuania) according to the manufacturer’s protocol. then the recombinant gene was transformed in to E. coli Top10 Fʹ, colony-PCR was used for detecting the recombinant clones.Also digestion by BglII restriction enzyme (Fermentas, Lithuania) was used to confirm hbpA sequence received clones. This enzyme has one restriction site at nucleotide 56 located in hbpA gene. A similar experiment was done on clones receiving 16S rRNA sequence using restriction enzyme MboI (Fermentas, Lithuania). This enzyme has one restriction site at nucleotide 244 on 16S rRNA sequence. The confirmed recombinant plasmids named pTZ57R/T-hbpA and pTZ57R/T-16S, representing plasmids contained hbpA and 16S rRNA genes respectively.
3.5. SYBR Green Real Time PCR Experiment
All reactions were performed using an ABI 7500 Fast Real-Time Amplification system (ABI, USA) and SYBR® Premix Ex Taq™ II (TAKARA, Japan). Real-time PCR was performed in a final volume of 20 μL containing2 µL of A. actinomycetemcomitans or T. forsythensis genomic DNA (35 ng/µL) as template, 2 μL of 2X master mix, 0.4 M each of the forward and reverse primers (Table 2). The thermal profile consisted of a 10 min of Taq polymerase activation at 95°C by 40 cycles of PCR (denaturation 95°C 15sec, annealing and extension 60°C 1 min). During the primer extension of each cycle, the increased amount of fluorescence from the amplified DNA was recorded by using the SYBR Green optic channel set at a wave length of 495 nm. The initial threshold value was set to 0.22 fluorescent units. A negative control with PCR-grade water instead of template DNA was used in this test. Following amplification, to verify the correct product by its specific melting temperature (Tm), a melting curve analysis of the amplified DNA was performed at 54 to 95°C, the temperature increasing rate of 0.2°C/sec was considered in this test. Also the reaction products were held at 4 - 8°C until were subjected to further analyzes by electrophoresis in 2% agarose gel.
3.6. Determination of Sensitivity and Limit of Detection (LOD)
To determine the reaction sensitivity or minimum copy number of the target genes that are able to show a visible bond in the PCR or a detectable signal in SYBR green real time PCR related melt curve analysis was measured. To this purpose, 10-fold serial dilution of the pTZ57R/T-hbpA (1740 pg - 1.74 fg) and pTZ57R/T-16S (42 pg - 0.42 fg) was prepared. After performing triplicate PCR or SYBR green real time PCR with the mentioned dilutions, the last dilutions demonstrating amplification of the target genes were determined as the LOD of the corresponding assay. Finally, the DNA concentrations of the last dilutions were converted as the copy number of the respective gene (27). To develop the SYBR green real time PCR tests into quantitative assays and copy number determination of the hbpA and 16S rRNA genes, the standard curves were generated by plotting the Ct values against log DNA concentration and linear regression was calculated using the ABI 7500 Data Analysis software, version 2.0.1.
3.7. Specificity Evaluation of the Assays
To determine the specificity and investigate the probability of non-target genes replication in other microorganisms, the genome of control negative bacteria (Table 1) was studied by the designed assays. To ensure the presence of replicable DNA in the negative control bacterial genomes, the amplification of universal 16S rRNA gene was done and confirmed samples were used for experiments of characteristics determination (28 ). Table 2 shows universal primers used for this evaluation.
4. Results
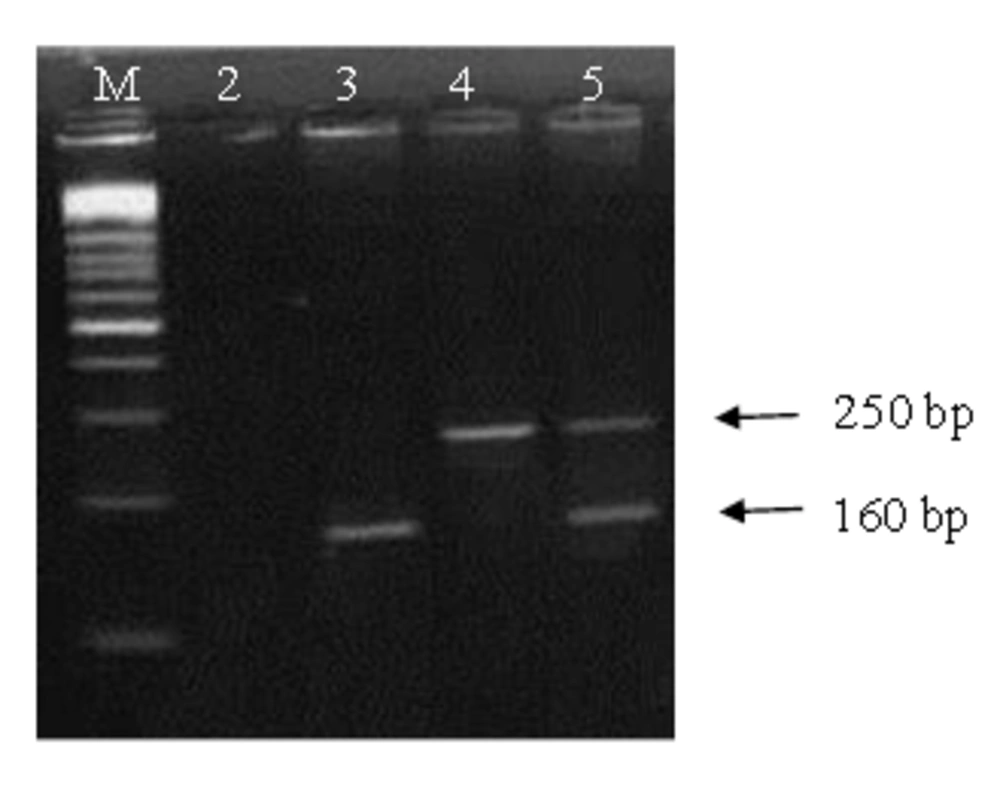
PCR assay results related to the two chromosomal genes, were positive for both hbpA and 16S rRNA genes. The optimum annealing temperature for amplification of the genes was 60.8°C. Amplified fragments of the hbpA and 16S rRNA with the lengths of 160 bp and 250 bp respectively were determined on the agarose gel. Gel agarose electrophoresis of the multiple assay products showed fast amplification of the both fragments (Figure 1). In SYBR Green real time PCR experiments, using the genes specific primers of A. actinomycetemcomitans or T. forsythensis , the related genomic DNA samples generated fluorescent signals. For the reactions using 100 ng genomic DNA of A. actinomycetemcomitans , a Ct value of 13 was obtained.
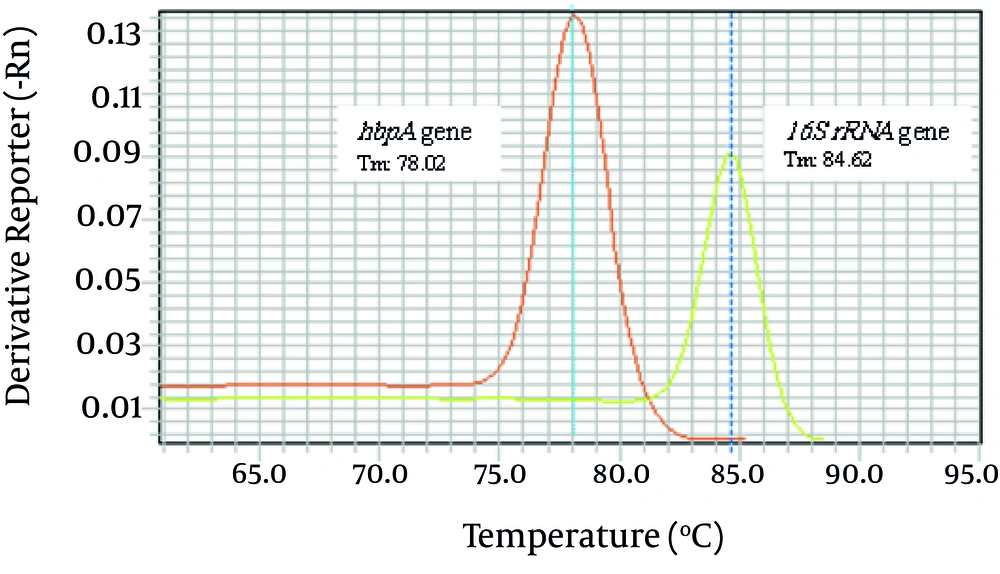
The Ct value of the reactions containing 100 ng genomic DNA of T. forsythensis was 12. Temperature melting analyses of the assays showed the Tm at 78.02°C and 84.62°C for hbpA and 16S rRNA genes respectively. Figure 2 shows the melting curve generated for detection of the genes using the ABI 7500 Fast real time amplification system. As expected agarose gel electrophoresis analysis of the products confirmed the amplification of the 160 bp and 250 bp bands (data not shown). No signal generation was seen in negative control tubes.
To prepare the sequences as positive controls, PCR products of hbpA and 16S rRNA genes were cloned in pTZ57R/T plasmid. Confirmation of the recombinant plasmids (pTZ57R/T-hbpA and pTZ57R/T-16S) conducted by PCR amplification of inserted sequences, and as expected, 160 and 250 bp bands on agarose gel were observed. Final confirmation of pTZ57R/T-hbpA plasmid by BglII restriction enzyme digestion resulted in a 3050 bp band on agarose gel. Also MboI restriction enzyme digestion of pTZ57R/T-16S plasmid resulted in a fragment with the length of 3136 bp on agarose gel (data not shown).
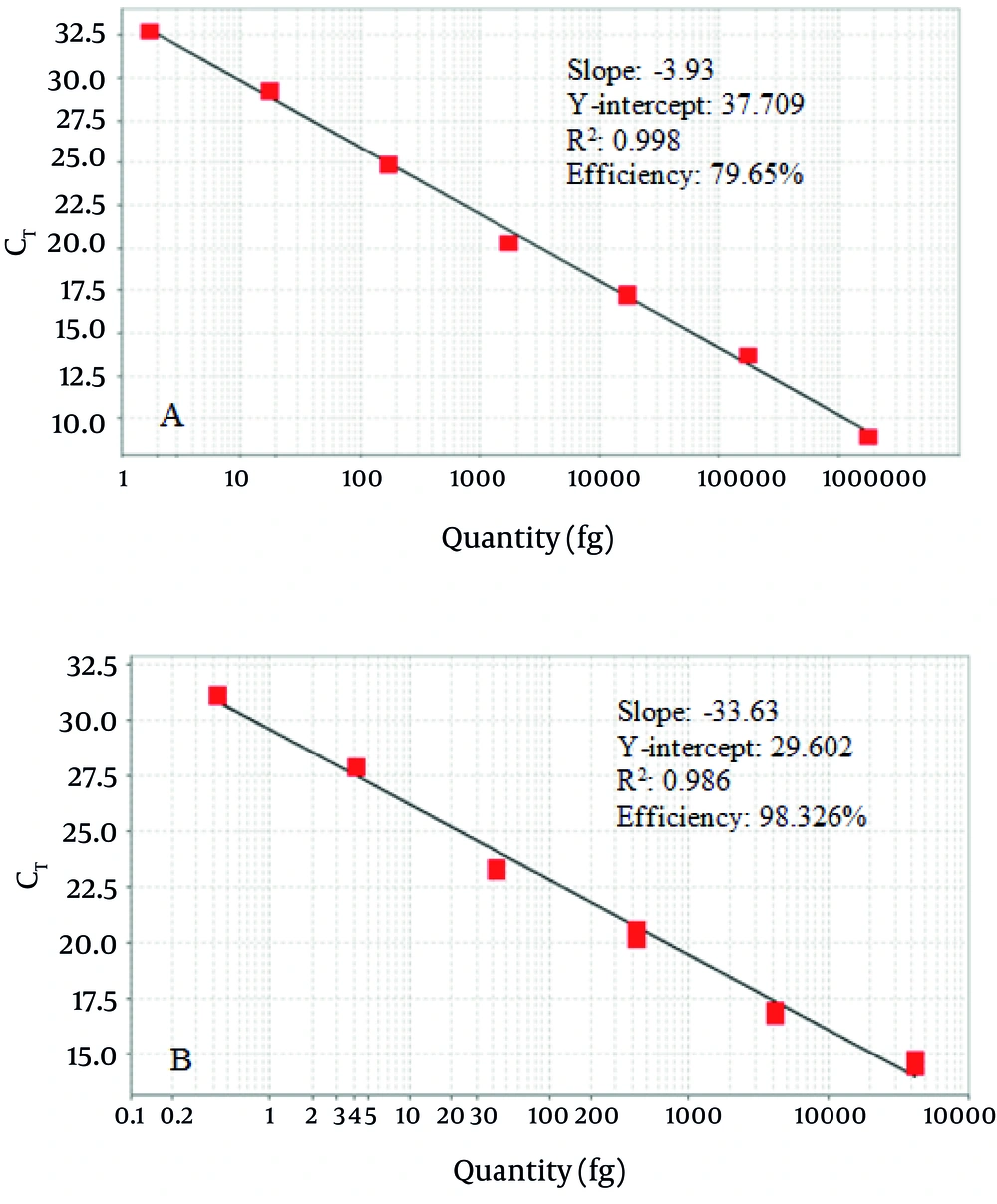
The last dilution of pTZ57R/T-hbpA plasmid that resulted in a distinct 160 bp band on agarose gel after conventional PCR was 17.4 fg. The last dilution for pTZ57R/T-16S plasmid was 4.2 fg (Figure 3, A1 and B1). Regarding the results, the lower detectable copy number for the hbpA and 16S rRNA genes of a 25 µL PCR reaction was considered as 5200 and 1200 copies respectively. After performing the SYBR Green real time PCR assays in triplicate on the 10-fold serial dilutions of pTZ57R/T-hbpA and pTZ57R/T-16S, the amplification curves confirmed the dilution increase and the standard curves were illustrated (Figure 3, A2 and B2). Comparing with the conventional PCR, the designed SYBR green PCR assays tenfold increased the test sensitivity. So the lower limit of detection of the assays for hbpA and 16S rRNA gene were 1.74 fg (520 copies) and 0.42 fg (120 copies) respectively.
The hbpA gene SYBR green assay was linear in the range of 1.74 - 1.74 ×10 6 pg of pTZ57R/T-hbpA (520 - 520 × 10 6 copies). The linearity for 16S rRNA gene SYBR green assay was in the range of 0.42 – 42 × 10 6 pg of pTZ57R/T-16S (120 – 120 × 10 5 copies). The slope of -3.93, Y-intercept of 37.709, R2 of 0.998 and amplification efficiency of 79.65% were the specifications of the regression line for the standard curve generated by the hbpA SYBR real time assay. The slope, Y-intercept, R2 and amplification efficiency of the regression line for the standard curve generated by the 16S rRNA SYBR real time assay was -33.63, 29.602, 0.986 and 98.326% respectively (Figure 4).
Conventional or SYBR real time PCR of both hbpA and 16S rRNA genes on genome of the negative control bacteria (Table 1) did not resulted to any band on agarose gel or fluorescent signal in the ABI 7500 Fast real time amplification system indicating the characteristics of both assays (data not shown). The PCR amplification of 16S rRNA universal gene on the genome of negative control bacteria resulted in a 475 bp band on agarose gel which indicated the presence of replicable fragment and the absence of PCR inhibitor (data not shown).
5. Discussion
Various studies in recent years have shown that oral diseases, such as periodontal diseases, are the risk factors or indicators for some systemic diseases such as coronary disease, stroke, peripheral vascular diseases, metabolic syndromes, hypertension, obesity and hyperlipidemia (29). Recently, it has been proposed that periodontal disease may be associated with breast cancer (30). Oral microbiologic profile determination could be a suitable approach for management of the oral diseases. Classical tests based on traditional phenotypic methods are very time consuming and inconvenient and on the other hand sometimes diagnosis and isolation of these bacteria are not easily done. In contrast, PCR as a steadfast, precise and sensitive method can be reasonable and suitable advanced replacement for diagnosis and control of these bacteria (7, 31).
In this study to achieve simple, rapid and accurate methods for detecting two of the most important bacteria in oral diseases, we designed and compared two diagnostic molecular assays. So a multiplex PCR assay and a quantitative SYBR green real time PCR for simultaneous detection of A. actinomycetemcomitans and T. forsythensis were designed and their analytical specificity and sensitivity was evaluated. One of the most important points in this approach is the primers design. After downloading sequences of the target genes from GeneBank of NCBI, they were aligned and then the consensus region was used for designing primers. It guarantees that the diagnostic primers anneal to all versions of the genes that are currently registered in GeneBank . It’s notable that in most of the studies, just one sequence has been used for primer design.
Multiplex PCR method is a suitable, precise and sensitive method due to reducing detection time with the basis of targeting different diagnostic genes of causative microorganisms . An additional important subject for designing multiplex primers is their Tm values which must be similar whereas the PCR products have different sizes. In this study compared with other similar studies, all the mentioned points were investigated. In a study by Leblebicioglu et al. in 2009, the PCR products were not differentiable and the band sizes were too short but the present study was designed in a way that the band sizes are at least 100 bp (32).
In this study, the PCR products were cloned into pTZ57R/T plasmid after optimized amplification of the target genes, and also to practice in following steps of the study the confirmed plasmids were used as positive controls. In nearly all other studies, the genomic DNA has been used as the positive control, Leblebicioglu et.al studies, in 2009 are the examples in this regard (32). It is obvious that cloning a gene into a plasmid vector in addition to inestimable access to the desired amount of amplified gene is possible to take advantage of having a stable positive control through cloning the gene into the plasmid.
The high sensitivity of the designed conventional PCR assay was determined via LOD determination. As a result, the LOD was 1200 copies for 16S rRNA gene and 5200 copies for hbpA gene. D'Ercole et al., in 2008 have compared the culture method and 16S rRNA gene multiplex PCR to detect the periodont pathogenic bacteria. The LOD of their multiplex PCR was 102 - 103 cells (33). Flemmig et al. in 1995, by used the artificial contamination method to calculate the LOD of leukotoxin gene lktA to detect A. actinomycetemcomitans finding 103 CFU/mL. In their study, enriching the samples on tryptic soy-serum-bacitracin-vancomycin agar prior to PCR improved the LOD to 102 CFU/mL (34). Conrads and his colleagues in 1999 determined the LOD of 16S rRNA PCR to detect T. forsythensis as 50 CFU/mL (35). The variation in the LOD estimations is partially due to the use of different primers and/or performing PCR using various thermal cycling programs. Also the selected methods for the estimation can influence the final results. LOD is one of the most important items in target amplification tests and there are various methods for its determination.
According the above studies one of these methods is preparing serial dilution of bacteria, counting and determining the CFU. Despite the high accuracy of these techniques, being time consuming and necessity to use live bacteria are some of its disadvantages. The other method to determine LOD is to measure the genome concentration, prepare serial dilution, estimate genome copy number in each dilution, conducting PCR at different dilutions, and finally estimate the LOD. However, one of the important disadvantages of this method is its low precision. According to various studies, cloning of target gene in plasmid and then preparing serial dilutions from it and PCR is one of the best methods for LOD determination (36) which was also used in the present study.
To improve the sensitivity or LOD of the conventional PCR assay and obtain a quantitative assay, we tried the same primers in SYBR green real time PCR approach. Fortunately, the hbpA- 16S rRNA SYBR green PCR assays at least tenfold increased the test sensitivity. Most of the oral pathogens are present in normal oral cavity with a low profile, but in periodontal diseases increase in their numbers can be happened. So a quantitative molecular method can play a critical role in evaluation of the health or disease of the oral cavity. Although quantitative anaerobic culture is a gold standard for number estimation of the oral pathogens, time consuming, laboring and relatively low level of sensitivity are some of its difficulties.
Various studies have shown a high concordance between results of anaerobic culture and real time PCR method in microbiological evaluation of periodontitis cases (25, 37-39). In our quantitative real time PCR experiments, the generated standard curves showed a linear range of 1.74 - 1.74 × 106 pg (520 - 520 × 106 copies) and 0.42 - 42×106 pg (120 – 120 × 105 copies) for pTZ57R/T-hbpA and pTZ57R/T-16S templates respectively. So to quantify unknown samples in the above ranges, the Ct value can be used on the standard curves of the respect target gene.to prove the claim, evaluation of the designed assays by using sample plaques of patients or artificially contaminated dental plaques is necessary.
The periodontopathogenic bacteria play an important role in the development and progress of the oral diseases. Thus, diagnosis of A. actinomycetemcomitans and T. forsythensis by using multiplex conventional and SYBR green real time PCR will supply necessary information about the oral bacterial profile and predicting disease in adult periodontitis. This study is the first report of developing and comparing conventional and real time multiplex PCR assay for the oral pathogens in Iran by designing primers and determining sensitivity and specificity, all the criteria necessary for a molecular diagnostic method are met. Domestication and design of the assays in Iran is an important step towards flourishing and development of medical diagnostic laboratories techniques for accurate and precise diagnosis of the oral pathogens.