1. Background
Bovine viral diarrhea (BVD) is an economically important disease of cattle with a worldwide distribution. BVD is caused by bovine viral diarrhea virus (BVDV), which belongs to Pestivirus genus within the family Flaviviridae. The virus has a nonsegmented positive sense single stranded RNA genome (+ssRNA) with about 12.5 kb length and consisted of a single long open reading frame flanked by 5’ and 3’ nontranslated regions (1). BVDV has two biotypes: those that induce cytopathic effects on cultured cells (cytopathic viruses), and those that induce persistent infection in cells without obvious cytopathology (noncytopathic viruses) (2).
BVDV infections in cattle are associated with a variety of clinical manifestations such as abortion, fetal resorption, congenital malformation, and the deadly mucosal disease (MD) (2). Infection of fetuses aged 40 - 120 days by noncytopathic biotype of BVDV may cause fetal immunotolerance, resulting in the birth of immunotolerant persistently infected (PI) calves that continuously shed the virus (3). The economic impact of BVDV infections has led a number of European countries to start eradication or control programs (4, 5). Diagnosis of BVD relies on laboratory-based detection of the viral causing agent or virus specific antibodies. The most common laboratory method for this purpose is ELISA.
BVDV genome encodes a large polyprotein which processed co- and post-translationally by host- and virus-encoded proteases to form at least 12 individual proteins (6). The order of the cleavage products in the BVDV (NADL strain) polyprotein is NH2-Npro-C-Erns-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH (1, 6). C, Erns, E1 and E2 are viral structural proteins and the other proteins are nonstructural (NS) proteins of the virus. The most immunogenic proteins of BVDV (NADL strain) are ErnsE2 and nonstructural protein 3 (NS3) (7). NS3 is an 80 kDa (p 80) protein containing an N-terminal serine protease domain and a C-terminal RNA helicase (1, 8, 9). The production of NS3 is essential for pestivirus RNA replication and the efficiency of this protein is regulated by a cellular chaperone that determine the viral cytopathogenicity (10). The NS3 is highly conserved among Pestiviruses and induces a strong humoral immune response in cattle exposed to live BVDV either naturally or by vaccination (11).
Antibodies againstNS2-3 and NS3 do not neutralize infectivity. However, the NS2-3 and NS3 (but not the NS2) are strongly recognized by polyclonal convalescent antibodies that (12) can be detected readily by serological tests, NS3 is known as an important antigen in BVDV serology (13). Animals vaccinated with modified live vaccines also have a strong antibody response to NS2-3 and/or NS3 protein (11). Therefore, NS3 is a proper candidate antigen to detect antibodies against the virus in the sera of infected animals (14). Recombinant NS3 has been expressed and evaluated in different researches but a major problem of those expressions was that generally, this protein is insoluble and aggregates as inclusion bodies in recombinant bacterial cells. Thus, the expressed recombinant NS3 must be treated with urea to modify it a soluble protein for the convenient and effective protein purification (15-18). Here, we used a different prokaryotic plasmid vector, pMAL-c2X, to express a soluble form of the protein with a high level of expression. This plasmid vector encodes a fusion protein called maltose binding protein (MBP) which is soluble and can even solubilize fused recombinant protein (19, 20).
2. Objectives
The main objective of this study was to develop an efficient bacterial expression system for production of recombinant NS3 protein to design a local ELISA kit in the country in the future.
3. Materials and Methods
3.1. Virus and Cell
The NADL strain of BVDV maintained in Razi Vaccine and Serum Research Institute was used as a reference BVDV. The virus was propagated in bovine turbinate (BT) cells and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 5% horse serum. After widespread viral cytopathic effects, the cells were subjected to one round of freezing at -40°C and thawing at room temperature. The virus was then harvested and stored at -40°C until viral RNA extraction.
3.2. PCR Primers
To synthesize the complementary DNA (cDNA) and for PCR amplification of the NS3 gene, the complete sequence of NADL strain of BVDV, with the accession number NC_001461, was retrieved from GenBank and was used to design the necessary primers. Primers which could amplify the complete sequence of NS3 gene were designed by Primer3 software.
3.3. Viral RNA Extraction and RT-PCR Amplification of NS3 Gene
Genomic RNA was extracted from the BVDV, previously propagated in BT cell line, by TriPure isolation reagent according to the manufacturer’s protocol (Roche, Germany). Complementary DNA (cDNA) was constructed by Revert Aid reverse transcriptase enzyme (Fermentas) and NS3 reverse primer (5’ GCCGCTGCAGTCACAACCCGGTCACTTGCTT), at 42°C for 1 h. For the amplification of the synthesized cDNA, 5 µL of cDNA was added to 45 µL of PCR mix containing 5 U of Super-Taq DNA polymerase (Gefanavaran, Iran), 5 µL of 10X Super-Taq PCR buffer, 2.5 µL of 50 mM MgCl2, 1 µL of 10 mM dNTPs mix, 1 µL (50 pmol) of each of NS3 forward (5’ GCCGTCTAGAGGGCCTGCCGTGTGTAAG) and reverse primers and 3% DMSO as an enhancer was also added. The reaction was run under the following thermal cycling program; pre-denaturing at 94°C for 3 min; denaturing at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 3 min, (37 repeats); final extension at 72°C for 15 min. RT-PCR products were analyzed by electrophoresis on 1% agarose gel in TAE buffer containing ethidium bromide (0.5 μg/mL).
3.4. Cloning and Expression of the NS3 Encoding cDNA in Escherichia coli
The cDNA amplified fragment, previously purified by a commercial gel extraction kit (BioNeer, Korea), and the pMAL-c2X plasmid (New England Biolabs, USA) both were digested with XbaI and PstI restriction enzymes (Fermentas, The Netherlands) at 37°C for 2.5 h and were extracted from agarose gel using a commercial kit (BioNeer, Korea). Digested DNA fragment and plasmid vector underwent ligation process at 22°C for 2 h followed by an overnight incubation at 4°C using T4 DNA ligase (Fermentas, Netherland). The ligation product was transformed into E. coli cells (TG1 strain) using Chung transforming method (21).the plasmids were extracted from colonies grown on ampicillin embedded LB agar media and the presence of the NS3 insert was confirmed by double-digestion using XbaI and HindIII or single digestion with BamHI.
Colonies with recombinant plasmids were cultured in ampicillin embedded LB broth media containing 20 mM glucose, until the OD600 reached to 0.5. Thereafter, Isopropyl-beta-D-thiogalactopyranoside (IPTG) was added to the cultures to a final concentration of 0.3 mM. After 4 h incubation at 37°C, expression of the recombinant MBP-NS3 protein was examined by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). MBP (maltose binding protein) is a 42.5 kDa protein encoded by pMAL-c2X plasmid. Three recombinant plasmids that well-expressed the protein were submitted to Gene-Fanavaran Company (Iran) for sequencing of the cloned NS3 DNA fragment. After sequencing, a plasmid that had no amino acid changing mutation was transformed into the BL21 strain of E. coli for a more efficient expression.
3.5. Western Blotting
E. coli BL21 strain expressing MBP-NS3, besides bacteria expressing only the MBP were subjected to SDS-PAGE and protein bands were transferred to a nitrocellulose membrane, using semi-dry procedure at 12 V for 90 min. Membrane was then blocked by 5% horse serum in PBST (phosphate buffered saline plus 0.05% Tween), at room temperature for 2 h. Membrane was rinsed three times with PBST and incubated for 1 h with 1/100 dilution of a bovine serum, previously determined as a positive serum, for antibody against BVDV (22). Afterward, the membrane was washed three times with PBST and incubated for 1 h with 1/1000 dilution of a rabbit anti-bovine IgG conjugated to horseradish peroxidase (Sigma). Finally, after washing by PBST and PBS, reactions were detected with 4-chloro-1-naphthol and hydrogen peroxide (H2O2).
4. Results
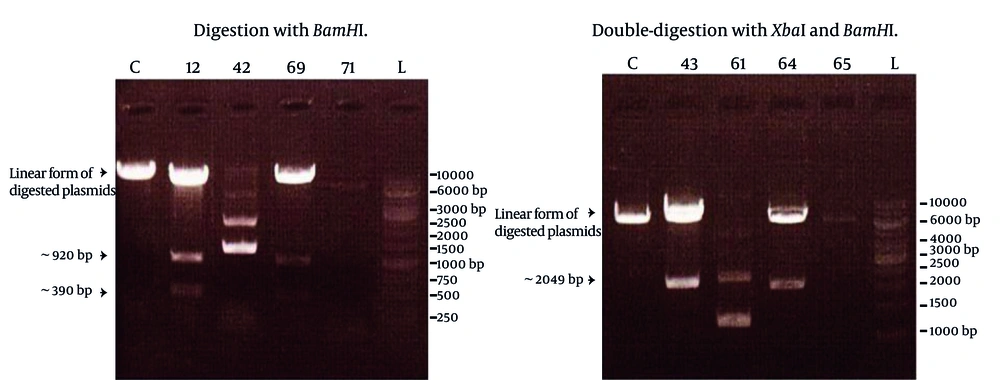
After observing cytopathic effects (most commonly, cell rounding, and detachment from the cell culture dish, and cell death) of the virus in the cell culture, total RNA was extracted from the cultivated infected cells. The primers designed in this study, successfully amplified a DNA fragment of about 2 kbp length which demonstrated the same expected length of NS3 gene. Amplified product was subsequently purified and digested with restriction enzymes XbaI and pstI , through the sites created by the forward and reverse primers, respectively. Digested PCR product was ligated into the pMAL-c2X plasmid, digested with similar enzymes, and transformed into the E. coli TG1 strain. Screening of eighty colonies appeared on LB agar supplemented with ampicillin led to the identification of four colonies carrying the recombinant plasmid. Screening was carried out using single digestion of the plasmids by BamHI enzyme or by double-digestion with XbaI and HindIII enzymes. In accordance with restriction enzyme map of NS3 and polylinker site of pMAL-c2X, digestion with BamHI produced a linear form of the plasmid plus two DNA fragments of about 390 and 920 bp length. Double digestion protocol definitely resulted in the production of two DNA fragments corresponding to the linear plasmid and NS3 (2049 bp) gene. The results of these digestions are shown in the Figure 1.
Lanes C represented control colony containing only pMAL-c2X, and the other lanes corresponded to four different colonies with their specified numbers. Lanes L represented a GeneRuler 1 Kb DNA ladder (Fermentas). Obviously, colonies 12, 69 (left) and 43, 64 (right) contained expected recombinant plasmid but colonies 42, 71 (left) and 61, 65 (right) were defective colonies.
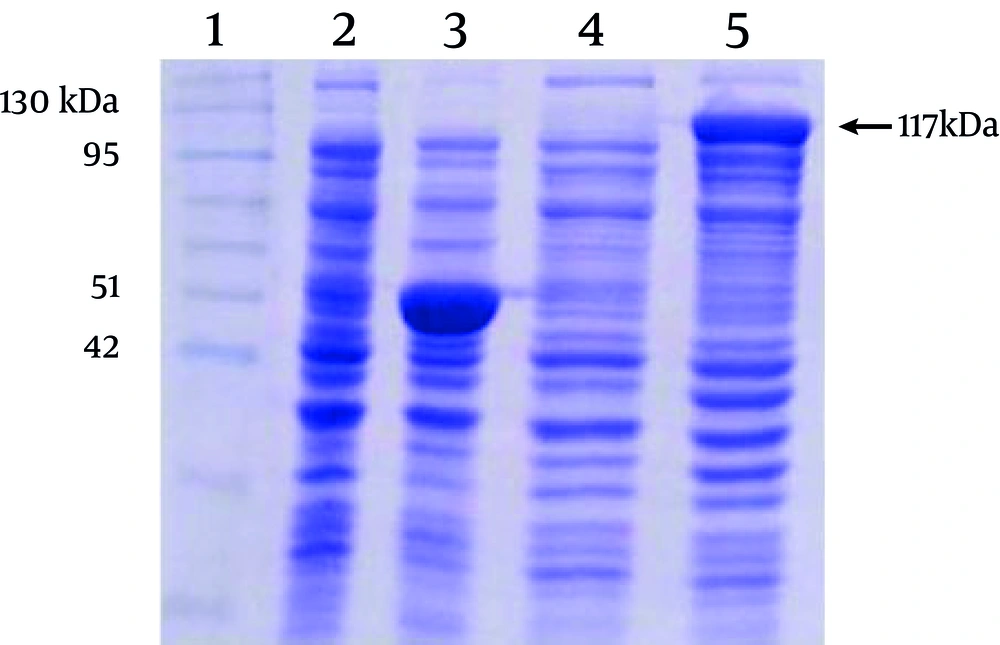
out of four colonies carrying recombinant plasmids, 12, 69, 43, and 64, only two colonies expressed MBP-NS3 protein, as determined by SDS-PAGE analysis. The results of the sequencing confirmed that in these colonies, plasmids contained the NS3 gene in a correct reading frame. Considering the molecular weight of MBP encoded by pMAL-c2X, predicted molecular weight of MBP-NS3 is about 117 kDa (MBP: 42 kDa and NS3:75 kDa). As shown in the Figure 2, expression of the recombinant MBP-NS3 is clearly detectable after adding IPTG to the culture of a recombinant bacterial colony.
Lane 1 indicates a pre stained protein molecular weight marker (Vivantis, Malaysia). Lanes 2 and 3 and lanes 4 and 5 represent the protein profiles of non-recombinant and recombinant bacteria before and after induction by IPTG, respectively. The expression of a protein of about 117 kDa, corresponding to the predicted molecular weight of MBP-NS3 is shown in the lane 5. The major protein of about 50 kDa, in the lane 3, corresponds to MBP plus α fragment of beta-galactosidase, expressed in non-recombinant bacteria after adding IPTG.
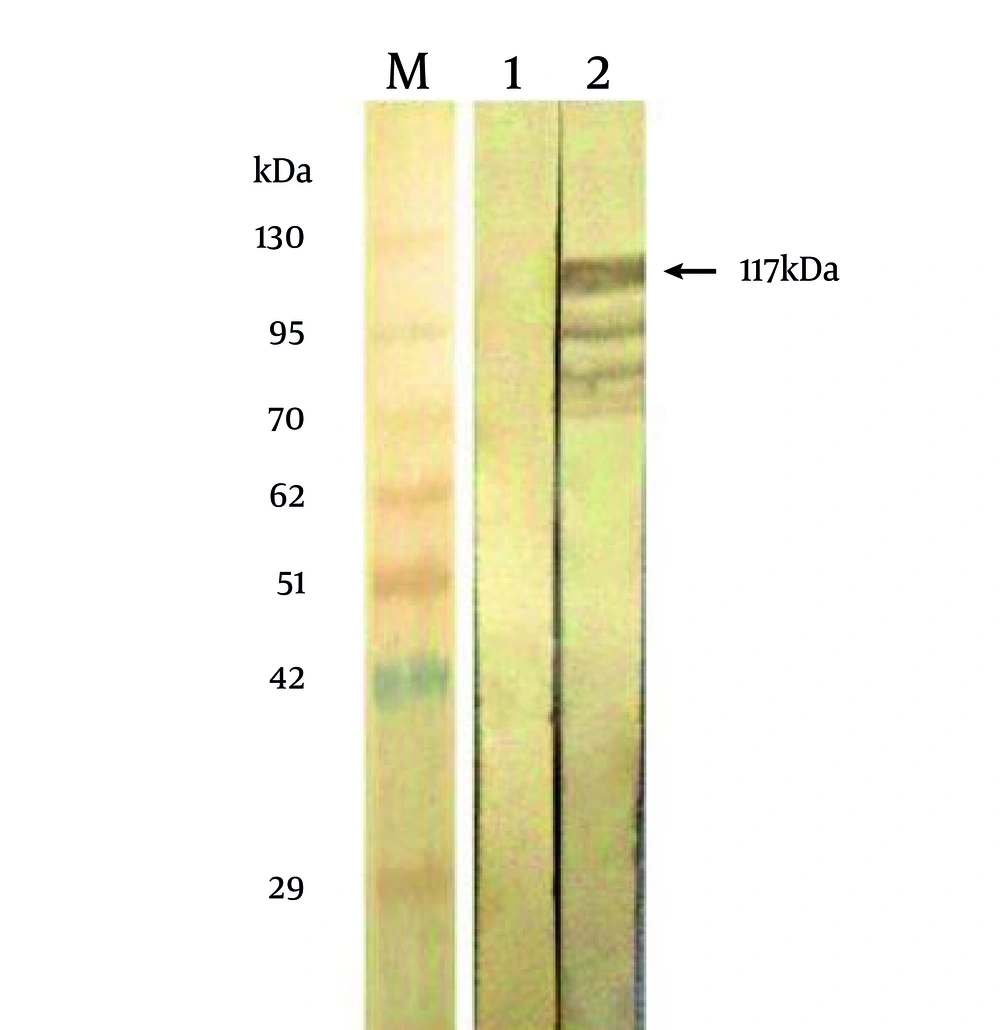
To confirm the identity of the 117 kDa protein, as being related to MBP-NS3, bacteria expressing this protein were tested by Western blotting, using bacteria expressing the MBP protein. The results showed (Figure 3) that the 117 kDa protein and some smaller bands reacted with a bovine serum, previously tested as an ELISA positive serum against BVDV. This pattern of the reaction is characteristic in majority of eukaryotic proteins which are expressed by E. coli . The appearance of the proteins bands smaller than NS3 indicates that a little quantity of NS3 is subjected to proteoloytic cleavage(s) in bacterial host cells. This finding was confirmed by the fact that there was not any reaction with the proteins of the non-recombinant bacteria.
5. Discussion
Bovine viral diarrhea virus nonstructural protein 3 has a critical role in the pathogenesis of bovine viral diarrhea disease of cattle. NS3 encoding gene is 2049 bp in length that encoded a protein consists of 683 amino acids (~80 kDa) containing an N-terminal serine protease domain and a C-terminal RNA helicase (6, 8, 9). Substitutions that eliminate serine protease activity abolish viral RNA replication, confirming its essential role in virus viability (23, 24). NS3 strongly induces humoral immune response in cattle exposed to live BVDV either naturally or by vaccination (11). This protein is highly conserved among Pestiviruses (25, 26). Donis and Dubovi (27) recorded high levels of antibodies to p80 from convalescent bovine sera.
Therefore, NS3 is one of the most important viral candidate proteins for detection of BVDV infection in cattle. Although, the extracts of virus infected cell cultures were formerly used in diagnostic ELISA kits as the detector antigens (28, 29), several ELISAs have recently been developed to detect BVDV infections using recombinant NS3 protein (7, 16-18). Eukaryotic or prokaryotic recombinant NS3 may be used for this propose and this protein has shown high sensitivity and specificity for detection of BVDV infection in comparison with whole virus antigen (16, 30). Hence, production of a recombinant form of NS3 in large amounts is economically very essential and useful for constructing BVDV antibody ELISA kits. Although it has been shown that eukaryotic expression of NS3 increases the sensitivity and specificity of ELISA kits (18), prokaryotic expression of NS3 is still considered because it is simpler and less expensive than eukaryotic expression system.
Prokaryotic expression of NS3 has been reported by several studies. Reddy et al. (17) cloned a 917-bp segment of NS3 into pGex-2T plasmid vector contained the glutathione-S-transferase (GST) gene. An 1152-bp cDNA fragment of NS3 (2/3rd of NS3 gene from C-terminus) was cloned into pGEMT Easy Vector by Bhatia et al. (31) and used as a detector antigen in their developed ELISA kits. Lecomte et al. (16) used a recombinant 80 kDa antigen of the BVDV/Osloss virus strain as a fusion protein with β-galactosidase for ELISA detection of BVDV specific antibodies. Vanderheijden et al. (18) inserted a 2183-nucleotide fragment of p80 sequence (Osloss strain) into the pARHS3 plasmid and analyzed expressed protein by ELISA.
Deregt et al. (15) carried out another study and cloned different regions of the NS3 gene (~500 – 900 nucleotides) of the type 1a Singer strain into the pET-30a-c (+) vector for expression of N-terminal six histidine-tagged constructs to map antigenic domains on the NS3 protein. A major problem of these prokaryotic expressions of the recombinant NS3 protein was that generally, this protein is insoluble and aggregates as inclusion bodies in recombinant bacterial cells. Therefore, the researchers had to use high concentration of urea to solubilize insoluble recombinant NS3 protein (inclusion bodies) and then try to renature it by gradient dialysis of the solution. Obviously, it is excessively time-consuming and expensive to follow theseprocesses.
Here, we cloned the encoding cDNA fragment of NS3 and constructed a recombinant plasmid vector which successfully expressed the recombinant NS3 using a prokaryotic expression system (E. coli BL21 strain). The reactivity of the protein with antibodies against BVDV was confirmed by western blotting. Although recombinant NS3 or its subunits have been previously expressed in prokaryotic systems, the present study describes expression of the protein in E. coli with a different vector, pMAL-c2X. The strong lac promoter of pMAL-c2X vector allows a high level of protein expression and the maltose binding protein tag, which is added to the N terminus of the molecule, facilitates the purification of the recombinant protein by Amylose resin. Several studies have shown that MBP is "a" soluble protein and can even solubilize recombinant protein expressed with as a fusion protein (19, 20).
Meanwhile, it has been shown that for the purification of the MBP fused recombinant proteins, rice flour can be used instead of Amylose resin. However, the efficiency of purification by rice flour is less than Amylose resin, but the extent of purification is sufficient to obtain a quantity of protein required for research purposes and moreover, it is much cheaper than amylose resin (32). Prokaryotic proteins expression with pMAL-c2X plasmid is one of the most efficient expression systems and by exploiting this system; we could produce a large amount of recombinant NS3 to develop BVDV specific ELISAs in Iran, in the near future. Developing such ELISA kits with more sensitivity, specificity, simplicity and less expensive than available commercial ELISA kits can improve detection of BVDV infections, help to eliminate the disease from herds, and decrease economic losses caused by this disease.