1. Background
The recent global spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases (ESBL) is a major concern because of new microbiological and epidemiological features.
Plasmid-mediated extended-spectrum β-lactamases extended spectrum beta-lactamase (ESBLs) are increasingly frequent among clinical isolates of the family Enterobacteriaceae throughout the world. The development of new enzyme groups that have a typical ESBL resistance phenotype but are non-TEM and non-SHV derivatives have recently been reported (1). CTX-M-type-lactamases constitute a novel group of enzymes encoded by Transferable plasmids (1). The CTX-M-type enzymes are a group of molecular class A extended-spectrum β- lactamases(ESBLs)that exhibit a general preference Tocefotaxime (CTX; hence the CTX-M name) and Ceftriaxoneand are capable to hydrolyze broad-spectrum Cephalosporins and are inhibited by Clavulanic acid, Sulbactam and Tazobactam. These enzymes are emerging in members of the family Enterobacteriaceae, that can cause resistance to CTX and other expanded-spectrum-lactams (2).
The first two CTX-M-type enzymes were reported in European countries in 1989 (1, 3). To this point, more than 40CTX-M-type-lactamases have been identified in various clinical isolates, mostly in Enterobacterial species such as Escherichia coli, Klebsiella pneumonia and Salmonella enterica serovar Typhimurium. These enzymes have been classified in to five major phylogenetic branches based on their amino acid sequence homologies (4).
The CTX-M group I including CTX-M-1,-3,-10,-12 ,-15,-28,-30 (5): and FEC-1; the CTX-M group II including CTX-M-2,-4,-5,-6,-7,-20 and Toho-1;the CTX-M group III including CTX-M-8; the CTX-M group IV including CTX-M-9,-13,-14(also named CTX-M-18),-16,-17,-19,-21,-24,-27 and Toho-2;and the CTX-M group V with CTX-M-25 and CTX-M-26. The CTX-M group consist of five groups , each group contains different types. Beta-Lactamases of the CTX-M group II are structurally related to the naturally produced beta-lactamase of Kluyvera ascorbata (6), and in CTX-M group III, is related to beta-lactamase of K. georgiana (7), and CTX-M group I enzymes are related to the beta-lactamases of K. cryocrescens (8). Although an identical enzyme to CTX-M-3 was isolated from K.ascorbata (9). The CTX-M group IV is related to enzymes from Kluyvera spp. isolated in Guyana, which was identical to CTX-M-14 (10).
CTX-M-producing strains were initially found in west Europe, they have now been observed over a wide geographical area, including Latin America(4), Africa, Asia (11), some parts of eastern Europe (12), and, recently, North America (13).
2. Objectives
In this work we report the dissemination of various CTX-M-Type beta-lactamases (CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9) in clinical isolates of Enterobacteriaceae from Arak educational hospital of University of Medical Sciences, Iran.
This study provides evidences about the global dissemination of CTX-M type ESBLs and emphasizes on the need of its epidemiological monitoring.
3. Materials and Methods
3.1. Bacterial Strains
350 randomly selected Enterobacteriaceae were isolated, during a 10-month period (May to February 2010), from clinical specimens Arak educational hospitals of Medical University, Iran. These organisms were screened for the presence of ESBL and then investigated for the presence of CTX-M-Type beta-lactamese. The isolated bacteria were kept at -70˚C before tested.
3.2. Phenotypic Detection of ESBL
ESBL producers were detected by combination disk methods (CLSI) (14). CAZ/CA (10 µg of clavulanic acid and 30 µg of CAZ) and CTX/CA (10 µg of CA and 30µg of CTX) (provided by Mast-German) disks were placed on the inoculated plates containing Muller Hinton agar.A positive test result was defined as a ≥ 5 mm increase in diameter of inhibition zone compared to a disk without Clavulanic acid (15).
Antibiotic susceptibility tests and Minimum Inhibitory Concentration (MIC) of antimicrobial agents that are usually active against the Enterobactericaeae were determined by an antibiotic disk diffusion method on Mueller-Hinton (MH) agar (Figure 1). Escherichia coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as controls.
MICs of Ceftazidime and Cefotaxime were determined by a micro dilution test using Cation-Adjusted Mueller-Hinton (MH) broth, in accordance with the Criteria of the Clinical and Laboratory Standards Institute (15). The MIC panels were prepared in house. Plates were incubated at 35°C for 18 hours before reading the results. E.coli ATCC25922 was used as a reference for quality control of in vitro susceptibility testing.
3.3. DNA Extraction and PCR Experiments
Genomic DNA template preparation using boiling methods and PCR amplification for CTX-M beta-lactamase genes were as follow: denaturation at 95 o C for 5 minutes, followed by 30 cycles of denaturation at 95 o C for 1minute, annealing for 1minute (according to Table 1), extension at 72 o C for 1 minute and final extension at 72 o C for 5 minutes. The primers, and fragment size are listed in Table 1.
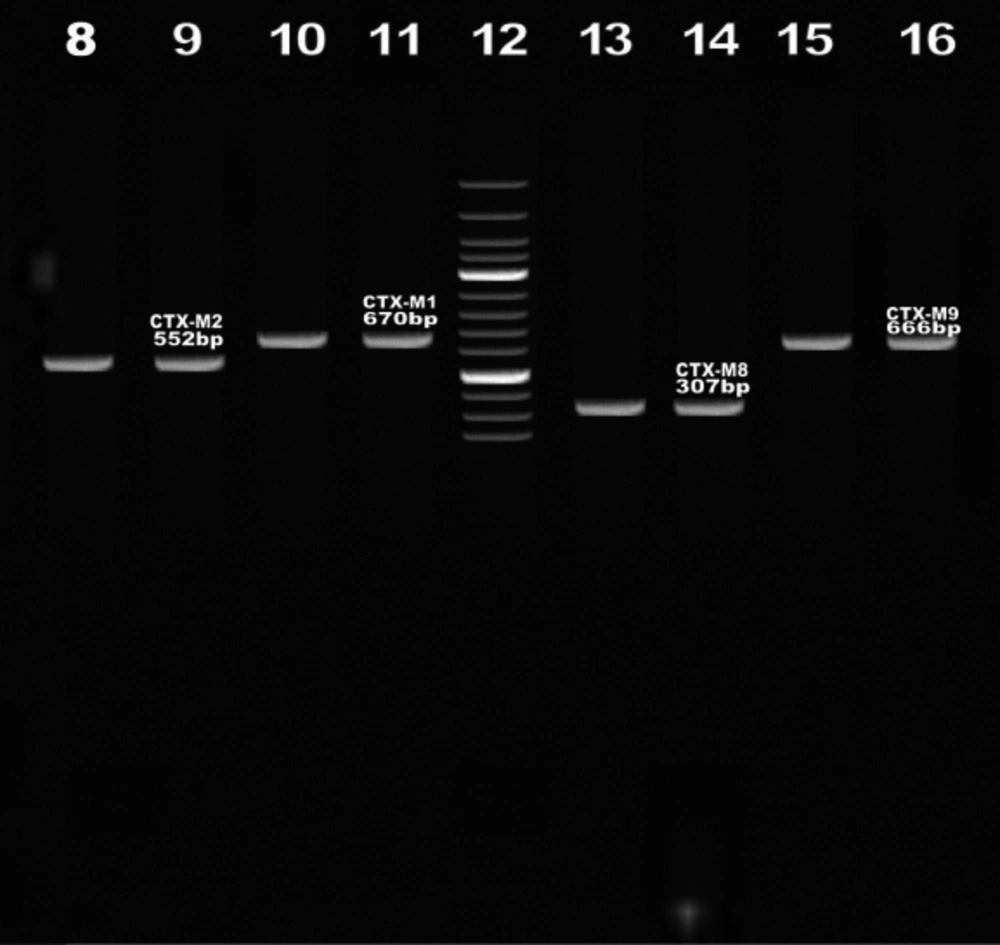
PCR products were subjected to electrophoresis on horizontal 1% agarose gels in TBE 1X buffer, loaded with 5 µL of reaction mix and stained with ethidium bromide after electrophoresis (Figure 2).
| Gene | Primer Sequence | Product Size, bp | Annealing Temp, °C | MgCl2 Concentration, mM |
|---|---|---|---|---|
| blaCTX-M1 | F: 5' AAGACTGGGTGTGGCATTGA 3' | 670 | 52 | 2 |
| R: 5' AGGCTGGGTGAAGTAAGTGA 3 | ||||
| blaCTX-M2 | F: 5'CGACGCTACCCCTGCTATT 3' | 552 | 60 | 3 |
| R: 5' CCAGCGTCAGATTTTTCAGG3' | ||||
| blaCTX-M8 | F: 5' CGC TTT GCC ATG TGC AGC ACC3' | 307 | 58 | 2 |
| R: 5' GCT CAG TAC GAT CGA GCC 3' | ||||
| blaCTX-M9 | F: 5' GCTTTATGCGCAGACGAGTG 3' | 666 | 50 | 2 |
| R: 5' GCCAGATCACCGCAATATCA 3' |
3.4. Statistics
Statistical analysis was performed using SPSS for windows version 11.5. Resistances were compared by Chi- square test. P value<0.05 was considered to be statistically significant.
4. Results
4.1. Bacterial Strains and Antibiotic Susceptibility Patterns
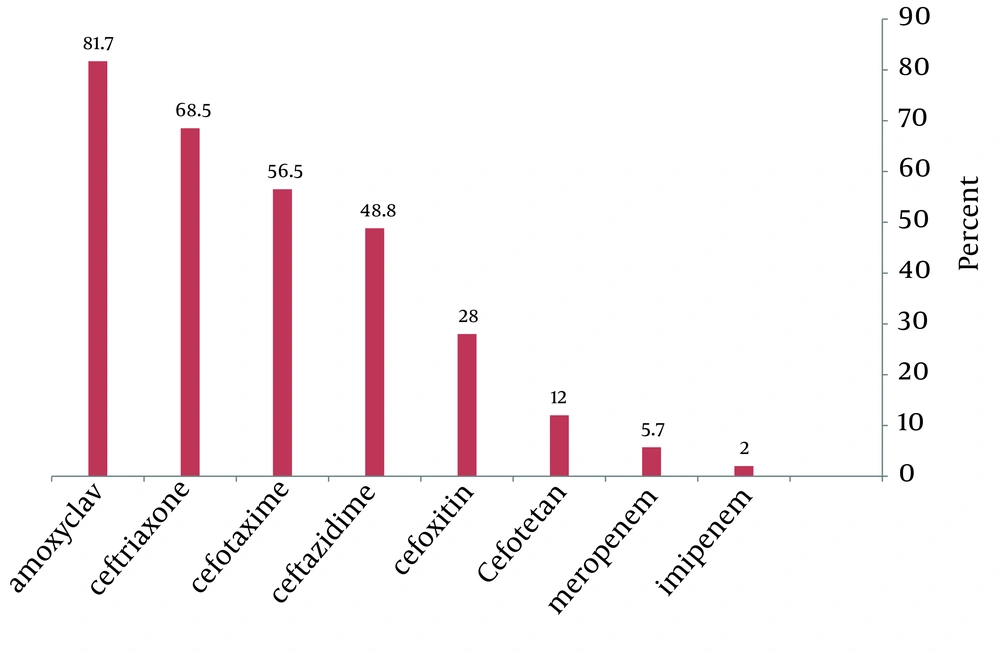
From May to February 2010, a total of 350 positive samples for Enterobacteriaceae were processed. 60% of the specimens were collected from hospitalized patients and 40% from outpatients urine (n= 251), wound (n= 42), blood (n=29), sputum (n= 28) samples. The resistance rate to the antimicrobial agents was as follows: Amoxyclav (81.7%), Ceftriaxone (68.5%), Cefotaxime (56.5%), Ceftazidime (48.8%), Cefoxitin (28%), Cefotetan (12%), Meropenem (5.7%), Imipenem (2%).
Among the tested third-generation Cephalosporins, the highest resistance was found against Ceftriaxone and Cefotaxime (68.5% and 56.5%, respectively).
Resistance phenotypes of ESBL- producing isolates were identified as follow: 80.5%(n=108) of isolates E. coli , 71.9% (n= 41) of isolates Klebsiella pneumoniae , and in 50% (n=5) of isolates Enterobacter cloacae, the ESBL producers were identified using the combined disk methods. The majority of ESBL producer that were mainly resistant to Cefotaxime and Ceftazidime, were obtained from urine (n=251) followed by wound (n=42), blood (n= 29), sputum (n= 28) specimens (Table 2). Also the most common ESBL positive phenotype was observed among hospitalized patients (48.5%).
| Sputum, % | Blood, % | Wound, % | Urine, % | |
|---|---|---|---|---|
| ESBL positive phenotype | 68.5 | 83.4 | 76 | 75.8 |
| ESBL negative phenotype | 31.5 | 16.6 | 24 | 24.2 |
4.2. MIC Determination
The MICs of the resistant isolates for Ceftazidime ranged between 16 and ≥512 µg/mL, whereas for Cefotaximethis amount varied between 64 and ≥ 512 µg/mL. According to our result more than 85%of the putative ESBL producers had MIC CAZ ≥ 16 and 98% of ESBL producer had MIC CTX ≥ 64. Nevertheless, a high diversity of the resistancelevel to Cefotaxime was observed for CTX-M-positive strains, as illustrated by the broad range of MICs (64 to ≥512 µg/mL) (Table 3).
| Isolates, µg/mL | ≤ 4 | 8 | 16 | 32 | 64 | 128 | 256 | ≥ 512 |
|---|---|---|---|---|---|---|---|---|
| Ceftazidime | 11 | 11 | 39 | 33 | 27 | 20 | 10 | 3 |
| Cefotaxime | 1 | 1 | 1 | 0 | 11 | 36 | 43 | 16 |
4.3. Detection of CTX-M-Producing Isolates
4.3.1. PCR Amplification
The resistant phenotypes of the 154 (44%) isolates were suggestive of CTX-M-type ESBL production that was obtained by Screening bla CTX-M determinants by PCR using four sets of Specific primers for the CTX-M family of ESBLs. Out of 154 ESBL positive, 92.2% isolates were CTX-M - 1, 28.5% isolates were CTX-M-2, 17.5% isolates were CTX-M-8, and 38.3% isolates were CTX-M-9 genes detected by PCR (Figure 2)(Table 4).
Also Table 5 shows the frequency of the beta-lactamase genesbased in isolated microorganisms from patients in different status.
| Isolates, % | CTX-M-Typebeta lactamase | ||
|---|---|---|---|
| Enterobacter cloacae | E. coli | K. Pneumonia | |
| 60 | 95.3 | 87.8 | CTX-M-1 |
| 0 | 35.1 | 14.6 | CTX-M-2 |
| 20 | 16.6 | 19.5 | CTX-M-8 |
| 20 | 45.3 | 21.9 | CTX-M-9 |
aP < 0.001
| Frequency | Hospitalization, % | Outpatients, % |
|---|---|---|
| CTX-M-1 | 91.1 | 49.2 |
| CTX-M-2 | 24.5 | 36.5 |
| CTX-M-8 | 8.8 | 34.6 |
| CTX-M-9 | 38.2 | 38.4 |
5. Discussion
There has been a dramatic increase in the number of CTX-M beta-lactamase producing organisms that were reported in the literature (5). This class of beta-lactamases has been recognized worldwide as an important mechanism of resistance to Oxyimino-cephalosporins used by Gram-negative pathogens.. In most of the cases, organisms producing these enzymes display higher levels of resistance to Cefotaxime and Ceftriaxone than Ceftazidime (5). However, phenotypic differentiation of organisms produce CTX-M beta-lactamases from organisms that produce other types of ESBLs can be difficult (12). Therefore, susceptibility testing which relies on identifying organisms that are resistant to Cefotaxime and/or Ceftriaxone but susceptible to Ceftazidime is not a reliable approach (5).
In our study antimicrobial susceptibility testing showed that the majority of isolates were resistant to at least one of the third-generation Cephalosporines. In a Study by Jeong et al. in 2004 in North Korea showed (16), Ceftazidime and Cefotaxime resistance was respectively 11% and 14%. Retrospective studies about resistance to antibiotics showed an increasing trend (17, 18).
Of totally 350 isolates, 203isolates (58%) showed ESBL phenotype detected by combination disk method which is different from the reported rates of ESBLs in other countries in our region such as India (97.1%), Turkey (57%) and Korea (68.7%) (19, 20).As 154(44%)of 203 Ceftazidime resistance or Cefotaxime resistance isolates were ESBL positive in this study, it appears that ESBL production has a significant role in resistance to Cephalosporines rather than other mechanisms of resistance such as the loss of porins and efflux pumps in our research (21).
In this study, the best coverage against ESBL-producing isolates was obtained with Imipenem. Although, the Carbapenem resistance has been rarely reported in past ,but its resistance rates have recently increased. Nonetheless , Carbapenem remains as the first choice for treatment of infections caused by ESBL-producing Entrobacteriaceae. It has been estimated that the worldwide Carbapenem resistance is near 2% (22). When national data are taken into account, this estimation are 0 to 8% (23) Most ESBLs caused resistance to one or more of the Oxyminobeta-lactams, the beta-lactamase does not always increase the MICs (24). The MICs of Ceftazidime in majority of ESBL positive isolates (n=132) were>16µg/mL.
Several studies revealed that distribution of ESBLs is widespread in our country. So the prevalence of E. coli and K. pneumoniae in 2009, 2010 in Iran were reported 52.5%, 59.2% (25, 26). in this study, the prevalence of ESBL producers among E. coli strains was 80.5%, the this amount among K. pneumonia strains was 71.9% and the among E. cloacae strains was 50%.
In this study, the frequency of CTX-M-1,-2,-8,-9 among isolated bacteria were 92.2%, 28.5%, 17.5%, 38.3% respectively. It has been reported that the presence of CTX-M-type beta-lactamases in Enterobacteriaceae isolates is up to 80% globally (27). In Turkey, recent studies reported that CTX-M-type beta-lactamases are very common in E. coli Isolated. It was reported that CTX-M-type beta-lactamase was obtained from 86.8% of community and hospital originated ESBL-producing strains (n= 61) (28). In another study, CTX-M-type beta-lactamase was detected in 13 (76.5%) of 17 ESBL-producing Enterobacteriaceae strains collecting from community (29). In a more recent study from Turkey, a high prevalence (98%) of CTX-Mtypebeta-lactamases was found in ESBL positive Enterobacteriaceae strains (n = 51) isolated from urinary tract infections (30).
Similar studies in Sudan, Korea, Thailand showed that the frequency of CTX-M-1 were respectively 45.9% ,16.4% ,14% compared with our findings is less abundant (31-33). Also, other studies showed the dissemination of CTX-M8 gene in Iran are higher than other parts of the world (34) On the other hand, in this study, the frequency of CTX-M-9 in isolates was less than Germany, America, Canada (ranged 38.5%-58.5%) (35, 36).
Today, an increase in bacterial resistance against antibiotics has become a major worldwide problem. During the past years, increasing rates of infections by extended spectrum beta-lactamase (ESBL)- producing isolates has greatly limited the use of non-carbapenem beta-lactam antibiotics, and thus the importance of Carbapenemand non-beta lactam antibiotics with therapeutic purposes has incrementally increased. The co-transmission of antibiotic resistance genes via plasmids may also affect the effectiveness of many antibiotics activities. Increased amount of antibiotic resistance rates among clinical isolates have resulted to higher morbidity and mortality and extended periods of hospitalization, consequently, has increased the economic costs (37).
From the results of this study, can be concluded that the number of ESBL – producing Enterobacteriaceae in patients enrolled in this study is higher compared with other parts of the world. Klebsiella species and E. coli are compromising the major microbial population. In order to resolve this problem, it is important to emphasize on the balanced and cyclic use of extended spectrum β-lactam drugs and imply an appropriate infection control strategy in hospitals. In addition, regular surveillance of resistance to antimicrobial agents is necessary.
Our findings were supported by the fact that many CTX-M cases were observed during hospitalization. These significant public health implications mean that the spread of bacteria producing ESBLs (particularly CTX-M enzymes) requires precise monitoring, enhanced surveillance and modifications have to be made in the antibiotic utilization policies with careful consideration, before the resistance predicaments worsen.