1. Background
Brucellosis is a bacterial disease caused by Brucella members and it is one of the important common diseases in human and animals and an important reason of abortion in animals (1-4). This disease is endemically found all around the world especially in Asian, African, North American and Mediterranean countries (5, 6). Annually so many people in the World are afflicted by this disease, hence, that is an important health problem in developing countries and its control in these areas is very important (5, 6). Several studies have shown that to control and prevent the disease, it is very important to have the epidemiological information. The microbiologists all over the world use various molecular and non-molecular methods to obtain this information (7).
The genus of Brucella includes several species and biovars. The difference between their species and biovars is mainly based on phenotypic characteristics of LPS antigens, sensitivity to colors, need to CO2, H2S production, metabolic features and phage typing (8, 9). But these methods are very difficult and their sensitivity is too low. On the other hand, since DNA homology for all species in the genus of Brucella is more than 90%, so the attempt is providing accurate identification of Brucella species and biovars, by molecular methods (8, 10).
Several molecular typing methods are introduced to find DNA polymorphism that is able to identify the Brucella species and biovars, among which detection of polymorphisms by PCR - RFLP has several advantages including the easy implementation, interpretation and use for large quantities of samples (11-13). In this method, by omp25, omp2 and omp31 loci and all Brucella species can be differentiated and their biovars identified (14). Several studies use these genes to differentiate Brucella species and biovars performed around the World (12, 15-17), but in Iran, no study was done especially on clinical samples by this method, and it is the first time that a study is done on molecular typing of B. melitensis and B. abortus isolated from blood samples.
2. Objectives
The aims of this study were Molecular typing and Molecular Epidemiology of B. melitensis and B. abortus from human blood samples based on PCR assay to find prevalence biovars in human blood.
3. Materials and Methods
3.1. Blood Samples Collection
160 Clinical specimens were received from patients who referred to Kerman clinical centers with Brucellosis detection as they had measurable antibody titers 1/80 during the last two years. 5 ml blood was taken from patients and the blood samples were transferred to laboratory and were kept in -20°C.
3.2. DNA Extraction
A modification of the method was described by Queipo-Ortuño et al. (18). Briefly, 0.5 ml of blood samples with 1 ml of erythrocyte lyses’ solution (320 mM saccharose, 5 mM MgCl2, 1% Triton X-100, 10 mM Tris HCl [pH 7.5]) were mixed and centrifuged at 15000×g for 2 min. The supernatant was discarded, and the above steps were repeated for four times until the pellet lost all reddish coloring. Four hundred microliters of nucleic lyses’ buffer(1% SDS ‚10 mM Tris-Hcl‚ 10mM EDTA‚ 10 mM Sodium Acetate [pH 8]) containing proteinase K (10 mg/ml) were mixed and incubated for 30 min at 55°C in shaker incubator. Then, 100 ml of ammonium acetate (7.5 M) was added and centrifuged at 15,000 ×g for 10 min. To the supernatant, two volumes of absolute ethanol were added, and after centrifuging at 15,000 ×g for 10 min, the pellets were dissolved in 25 µl of TE buffer (pH 8.0) and stored at 4°C for PCR or at -20°C for long-term storage.
3.3. Amplification of omp2a and omp2b Fragments
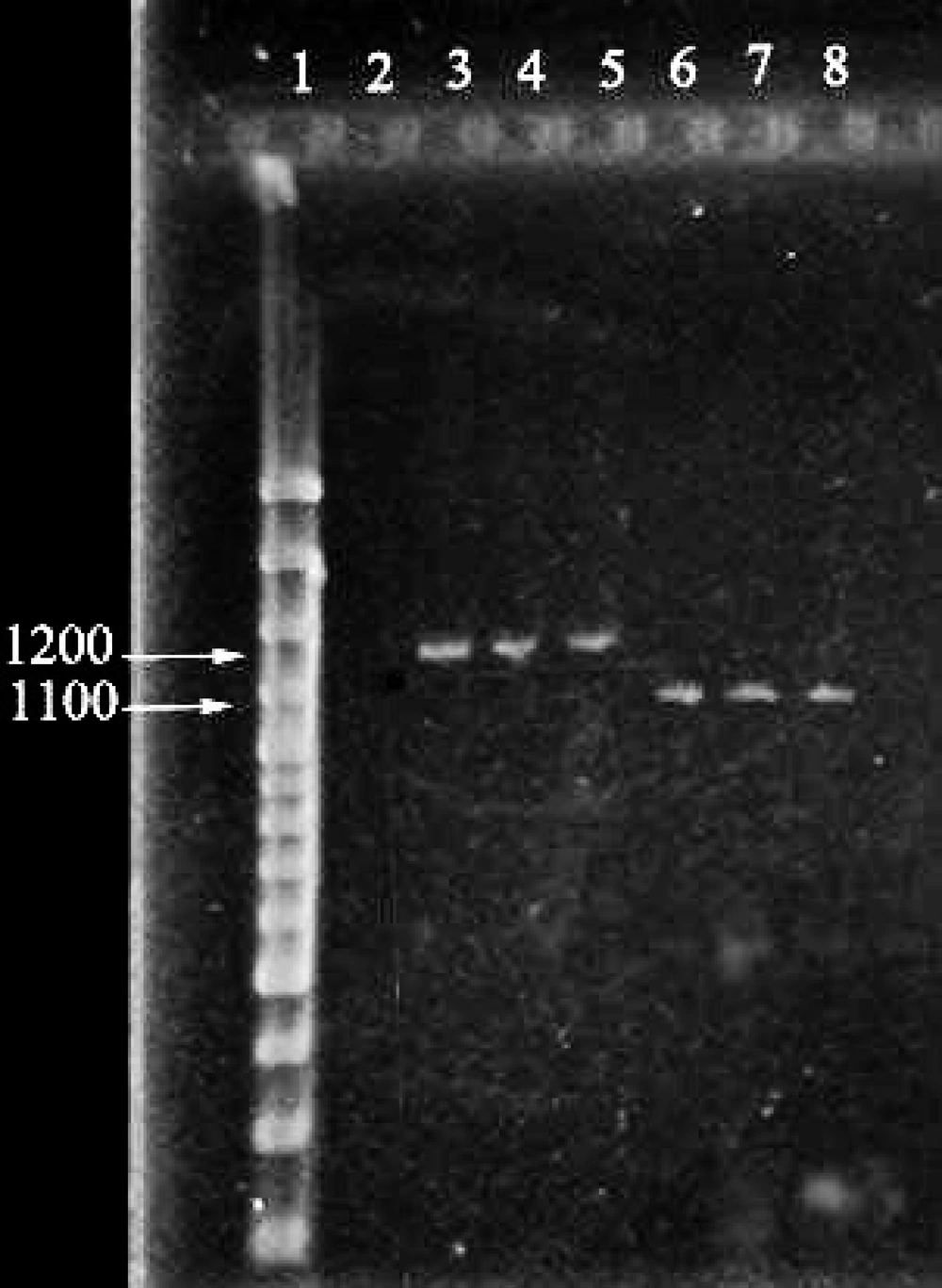
Cloackaert et al. ( 13 ) designed primers for amplification of omp2a and omp2b fragments and their sequences are shown in Table 1. Each PCR reaction mixture contained; 1X PCR buffer, 2 mM MgCl 2 , 1 μl template DNA (0.5 μg) , 0.15 mMdNTP, 2.5 U Taq DNA polymerase, 20 pmol of each forward and reverse primers and sterile distilled water up to 50 μl. According to the following program, PCR for omp2a was performed in a GenAmp PCR system (Eppendorf, Germany): pre-denaturation for 5 min at 94°C followed by 35 cycles each containing denaturation at 94°C for 60 sec, annealing at 50°C for 120 sec and extension at 72°C for 180 sec, followed by final extension at 72°C for 7 min. Also PCR for omp2b was performed according to the following program: Pre-denaturation for 5 min at 94°C followed by 35 cycles each containing denaturation at 94°C for 45 sec, annealing at 58°C for 60 sec and extension at 72°C for 60 sec, followed by final extension at 72°C for 7 min. Then, The PCR products were analyzed using the electrophoresis technique on 2% agarose gel for 1 hour at 25 mA, stained by SYBR-Green and visualized under UV transilluminator (Figure 1). Finally, amplification products were further evaluated restriction digestion procedures.
| Primer | Sequence |
|---|---|
| omp2a F | 5’-GGCTATTCAAAATTCTGGCG-3’ |
| omp2a R | 5’-ATCGATTCTCACGCTTTCGT-3’ |
| omp2b F | 5’-CCTTCAGCCAAATCAGAATG-3’ |
| omp2b R | 5’-GGTCAGCATAAAAAGCAAGC-3’ |
3.4. Enzymatic Digestion
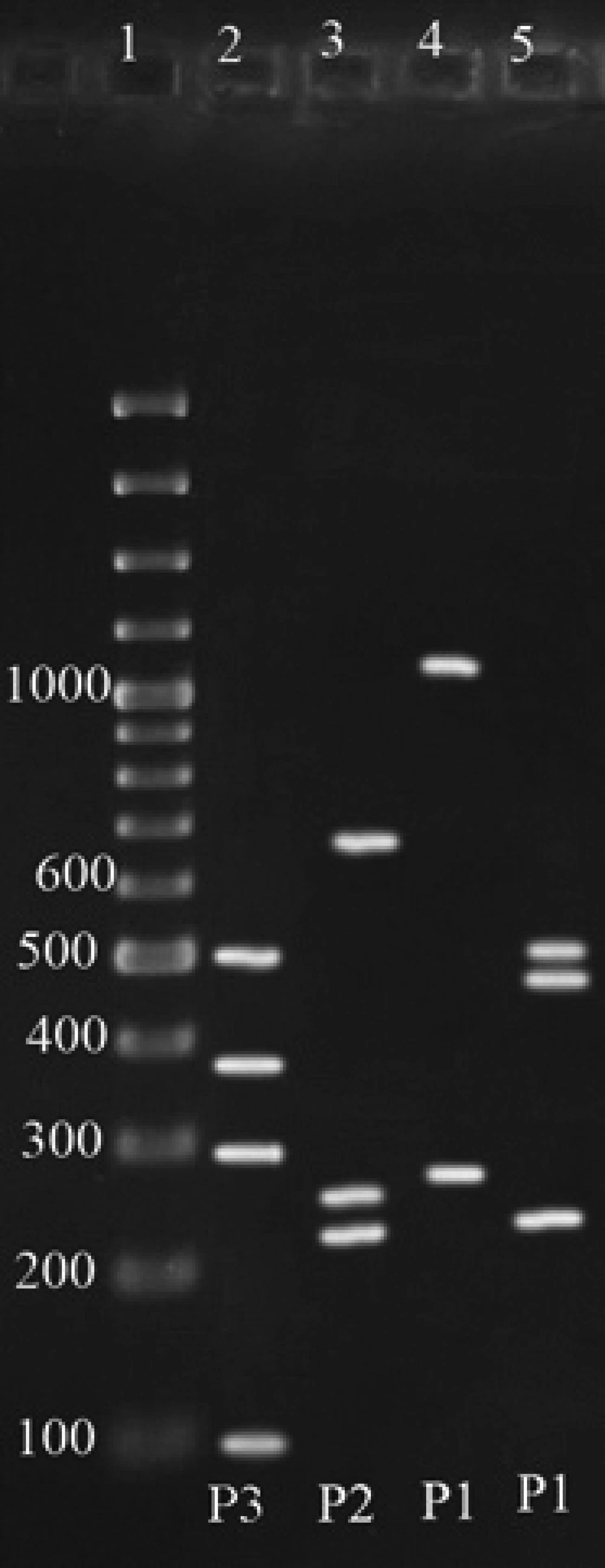
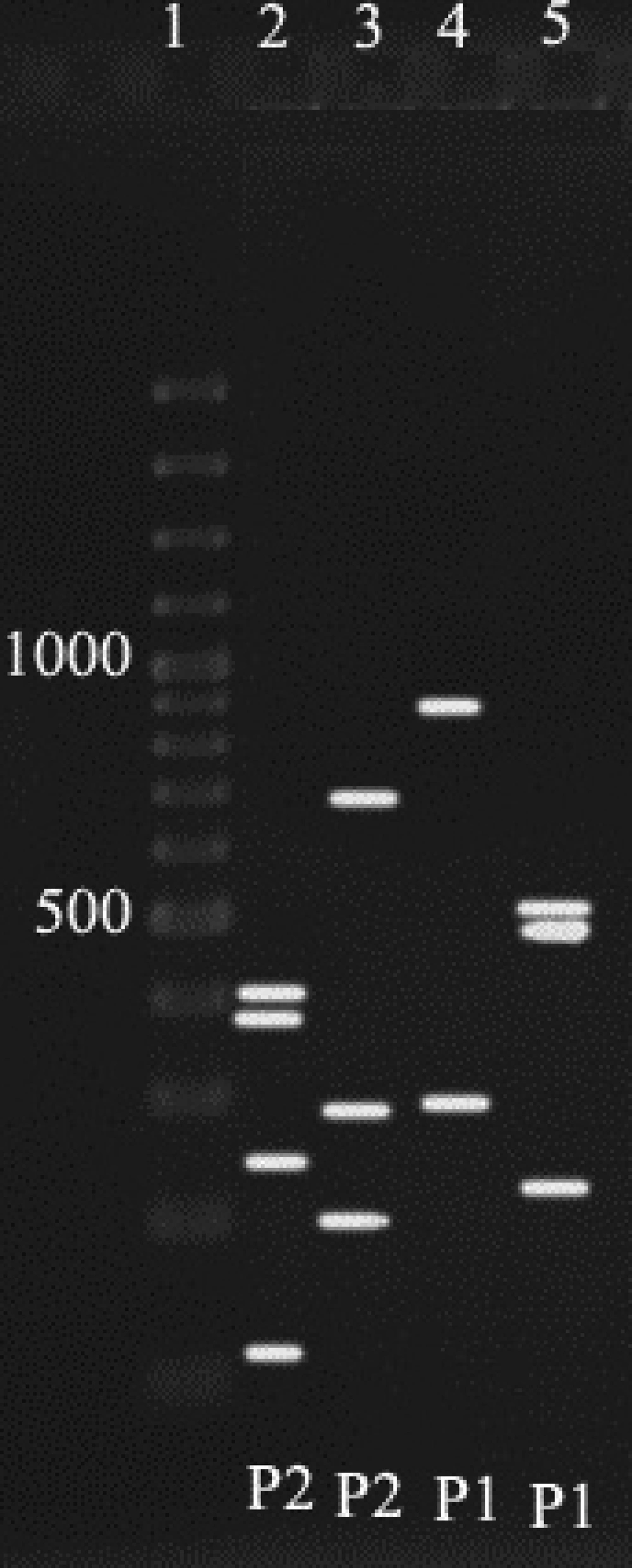
To identify polymorphisms, the amplified products were subject to restriction enzymes. For enzymatic digestion or PCR-RFLP, the following 15 μl reactions were prepared for positive samples. 6 μl free nuclease sterile water, 1.7 μl enzyme buffers, 0.3 μl restriction enzyme (10-20 U), and 7 μl of PCR product. After providing the above reaction, micro tubes were put in the water bath for 2 hrs at 37°C. After 2 hours, products of RFLP were electrophoresed on 2% gel agarose. Finally the size of the fragments which were resulted from enzymatic digestion of all positive PCR samples compared with each other and according to the results of Cloackaert et al. (15), the biovars were identified.
3.5. Statistical Analysis
Statistical analysis was conducted to determine how many samples were positive for each bacterium, as well as those positive for two bacterial species. Perspective analyses were performed and data rounded numerical values (percentage) were documented.
4. Results
In 160 blood samples studied by PCR technique, 52 cases obtained bands of 1100 bp foromp2alocus and 1200 bp for omp2b locus which indicates that the samples are positive for Brucella (Figure 1).
According to Cloackaert et al. ( 13 ) regarding enzymatic digestion of the amplified fragments, and comparing the results of the present study with their obtained patterns, for 52 cases from digestion of omp2a fragment by Pst1 and Hinf1 enzymes, P3 and P2 Patterns were obtained respectively, and for omp2b, P1 and P1 patterns were resulted, which indicates the B. melitensis biovar 1 (Figure 2) ( 15 ),and for 33 cases (44.5%) from digestion of omp2a fragment by Pst1 and Hinf1 enzymes, Patterns of P2 and P2 and for omp2b patterns of P1 and P1 were resulted respectively, which indicates the B. abortus one of the biovars 3, 5, 6 or 9 (Figure 3). As a final conclusion, the used primers and enzymes in the current study showed that they were able to differentiate between B. melitensis biovars, but they cannot separate all of the B. abortus biovars from each other.
5. Discussion
Due to differences in the pathogenicity of Brucella species and biovars, with a view to epidemiology of brucellosis, recognition of Brucella biovars is important and hence typing of various strains is the main task of control centers of brucellosis and must be performed continuously (9, 13, 15). Distinction between the Brucella species and biovars in the past was mainly based on phenotypic characteristics. Today, with advancement of molecular techniques, several molecular typing methods to find DNA polymorphism are used, which are capable to identify Brucella biovars and species. The PBR assay is a technique used for typing (9, 10, 19). The current study used this technique to determine the species and biovars of B. abortus and B. melitensis isolated from clinical specimens in Iran.
Different genetic fragments such as omp31, 16srDNA, omp2 and so on are used in different studies, to identify and differentiate Brucella species and biovars, which could not show ability to differentiate Brucella biovars completely (14, 20, 21). Therefore, in subsequent studies especially in Cloackaert et al. study, omp2a and omp2b fragments and several restriction enzymes were used (15). These primers and enzymes were used in different parts of the world by different people, also in central parts of Iran they were used by Salehi et al. for molecular typing of common Brucella biovars (15-17, 22).
Results of all these studies, like those of the current study showed that the primers and enzymes are able to differentiate B. melitensis biovars but could not distinguish all of the B. abortus biovars from each other. However, present study differs from most of the studies in type of sample; in the current study human clinical samples were used whereas the others used animal samples. Also compared with the studies in Iran; there were differences, such as sample types, the enzymes used, and the sample figure. Salehi et al. by one or two pairs of primers and an enzyme identified Brucella isolated from animal samples (22). But in the current study, isolated Brucella from clinical samples were typed with two pairs of primers and two restriction enzymes. They were not able to differentiate between other B. melitensis biovars and biovars of B. abortus, but results of the current study showed that two species can be differentiated from each other and one can determine their biovars.
The results of this study like others in different parts of Iran showed that B. melitensis biovar 1 is the predominant form of these species in Iran (23, 24). Results of the current study showed that one of the B. abortus biovars 3, 5, 6 and 9 is the dominant form of these species, according to the previous studies in Iran, the biovar 3 was more likely (23, 24).
Same as the results of other studies in other parts of the world, the current study (15-17) showed that the PCR-RFLP techniques, by the primers and enzymes, were able to differentiate species and biovars of B. abortus and B. melitensis in clinical samples well. So they can be used to molecular typing of biovars isolated from different samples. Also comparing the obtained results with those of the other studies showed that the dominant pattern which causes the disease of brucellosis, especially B. melitensis, was not different in Iran, so, control and preventive measures of relevant organizations were appropriate and this trend should continue.