1. Background
The food microbial safety is a major concern of consumers, industries and regulatory agencies all over the world. The rapid and accurate identification of bacterial pathogens is important, both for quality assurance and to trace bacterial pathogens within the food supplies (1, 2). During the last few years, international standards have been agreed the use of PCR-based detection of food-borne pathogens and legislations are implementing new types of analyses as the accepted official methods. For example, European regulation EC 2073/2005 allows the use of alternative detection methods based on certified analyses of international standards (3). Multiplex PCR is a relatively new method that can simultaneously amplify template mixture and decrease the detection costs, conquering the weakness of single PCR detecting only one template once (2, 4, 5).
In studies of food-borne pathogens, three major pathogens have emerged as being of significant importance in terms of human health and disease. These include: Escherichia coli O157:H7, Listeria monocytogenes, and Salmonella spp. These organisms have frequently been associated with meat and meat products and linked to a number of cases of human infections (6-8).
One of the most significant food-borne pathogens that have gained increased attention in recent years is E. coli O157:H7. E. coli O157:H7 is a serious pathogen included in the verocytotoxigenic group of E. coli. This serotype has been found to be responsible for a wide range of illnesses in humans, including hemorrhagic colitis and hemolytic uremic syndrome. Although cattle have long been considered the principal reservoir of E. coli O157:H7, other domestic animals and their carcasses contain this microorganism (9-11).
L. monocytogenes is the causative agent of listeriosis, a disease that can be serious and is often fatal in high risk groups such as pregnant women, neonates, and immunocompromised adults with a mortality rate that may reach to 75% (12, 13). Different food types including raw milk, fish, beef, lamb, pork and chicken, have been implicated as the sources of contamination (14, 15).
Salmonella, a causative agent of gastroenteritis, has frequently been associated with poultry which are considered the primary source; however other meats such as beef, lamb, and pork have also been implicated as sources of contamination (15, 16). Typical symptoms of disease associated with salmonellosis including nausea, vomiting, and diarrhea; additional complications associated with infection include septicemia, or reactive arthritis (17).
2. Objectives
The present study was carried out to determine the prevalence of E. coli O157:H7, L. monocytogenes, and Salmonella spp. in beef, buffalo and lamb marketed in Ahvaz, South-west part of Iran using multiplex PCR technique.
3. Materials and Methods
3.1. Sampling and Enrichment Procedure
During a 10-month period, a total of 210 samples of beef, buffalo and lamb, each sample weighted approximately 500 g, were purchased from retail outlets and popular supermarkets. All samples were placed in cold portable insulated boxes, transported to the laboratory in ice and processed within 2 hours of collection.
The samples were minced or cut into pieces and three representative sub-samples (25 g each) were homogenized in a stomacher (Bagmixer 400W, Interscience, St. Nom, France) with 225 mL of enrichment broths. Enrichment of the pathogens was carried out using E. coli enrichment broth, Listeria enrichment broth and Rappaport-Vassiliadis enrichment broth for E. coli O157:H7, L. monocytogenes , and Salmonella , respectively (Table 1). Enrichment broths were incubated for 20-24 hours at 35ºC, except for Salmonella which was incubated at 42ºC.
| Enrichment Broths | Composition |
|---|---|
| Listeria enrichment broth | Peptone from soymeal (3 g/L), Peptone from casein (17 g/L), Yeast extract (5 g/L), D- Glucose (2.5 g/L), NaCl (5 g/L), KH2PO4 (2.5 g/L), Nalidixic acida (0.02 g/L), Polymyxina(0.01 g/L), Acriflavin hydrochloridea (0.015 g/L), Cycloheximide (0.05 g/L), pH 7.3 ± 0.2. |
| E. coli enrichment broth | Peptone from meat (10 g/L), NaCl (2.5 g/L), KH2PO4 (0.75 g/L), K2HPO4 (2 g/L), Lactose (2.5 g/L), Bile salts (0.66 g/L), Sodium novobiocina (0.01 g/L), pH 6.9 ± 0.2. |
| Rappaport-Vassiliadis enrichment broth | Peptone from soymeal (4.5 g/L), Magnesium chloride hexahydrate (28.5 g/L), NaCl (7.2 g/L), KH2PO4 (1.26 g/L), K2HPO4 (0.18 g/L), Malachite green oxalate (0.072 g/L), pH 5.2 ± 0.2. |
3.2. DNA Extraction and the Multiplex PCR Reaction
The DNA extraction was carried out through boiling (5). One mL sample of the enrichment broths were centrifuged at 14000 rpm for 3 minutes. Bacterial pellets were resuspended in 1 mL of sterile saline solution (0.85% NaCl). Following the centrifugation the supernatants were replaced with 50 µL of sterile distilled water and incubated at 100ºC for 10 minutes to release the bacterial DNA. After boiling, the clear supernatants obtained by 5 minutes centrifugation at 14000 rpm, were collected and stored at -20ºC until use.
The extracted DNA from each specific enrichment broth was mixed together and used in multiplex PCR reactions. The oligonucleotide primers used in this study (Table 2) and the multiplex PCR was carried out according to the procedure reported by Kim et al.(4). PCR reactions were performed in a thermal cycler (Eppendorf, Mastercycler Gradient) under the following conditions: initial denaturation at 94ºC for 3 minutes, denaturation at 94ºC for 45 seconds, primer annealing at 54ºC for 45 seconds, and extension at 72ºC for 60 seconds. The final cycle included a 5-minute additional extension at 72ºC. PCR products were observed following gel electrophoresis on 2% agarose gels using ethidium bromide staining.
| Bacterium | Primer Sequence (5´→ 3´) | Target Gene | Product Size (bp) |
|---|---|---|---|
| L monocytogenes | Forward:CTGGCACAAAATTACTTACAACGA | p60 protein | 454 |
| Reverse AACTACTGGAGCTGCTTGTTTTTC | |||
| E. coli O157:H7 | Forward:GATAGACTTTTCGACCCAACAAAG | Shiga-like toxin | 208 |
| Reverse:TTGCTCAATAATCAGACGAAGATG | |||
| Salmonella spp. | Forward:GAATCCTCAGTTTTTCAACGTTTC | invA | 678 |
| Reverse TAGCCGTAACAACCAATACAAATG |
3.3. Limits of Detection of the Multiplex PCR Reactions
The sensitivity of the multiplex PCR reactions was assessed on artificially contaminated beef, buffalo and lamb samples. Twenty-five grams of samples were homogenized in 225 mL of each specific enrichment broths and spiked with 1 mL of serial decimal dilutions of the specific pathogen. The bacterial concentration of the contaminating cultures was determined by culturing on TSA. For each bacterium and product, three levels of contamination were tested (Table 3). A non-spiked sample was used as a negative control. The homogenates were enriched as described in Section 2.1. DNA extraction and the multiplex PCR were performed in the same way as explained above.
4. Results
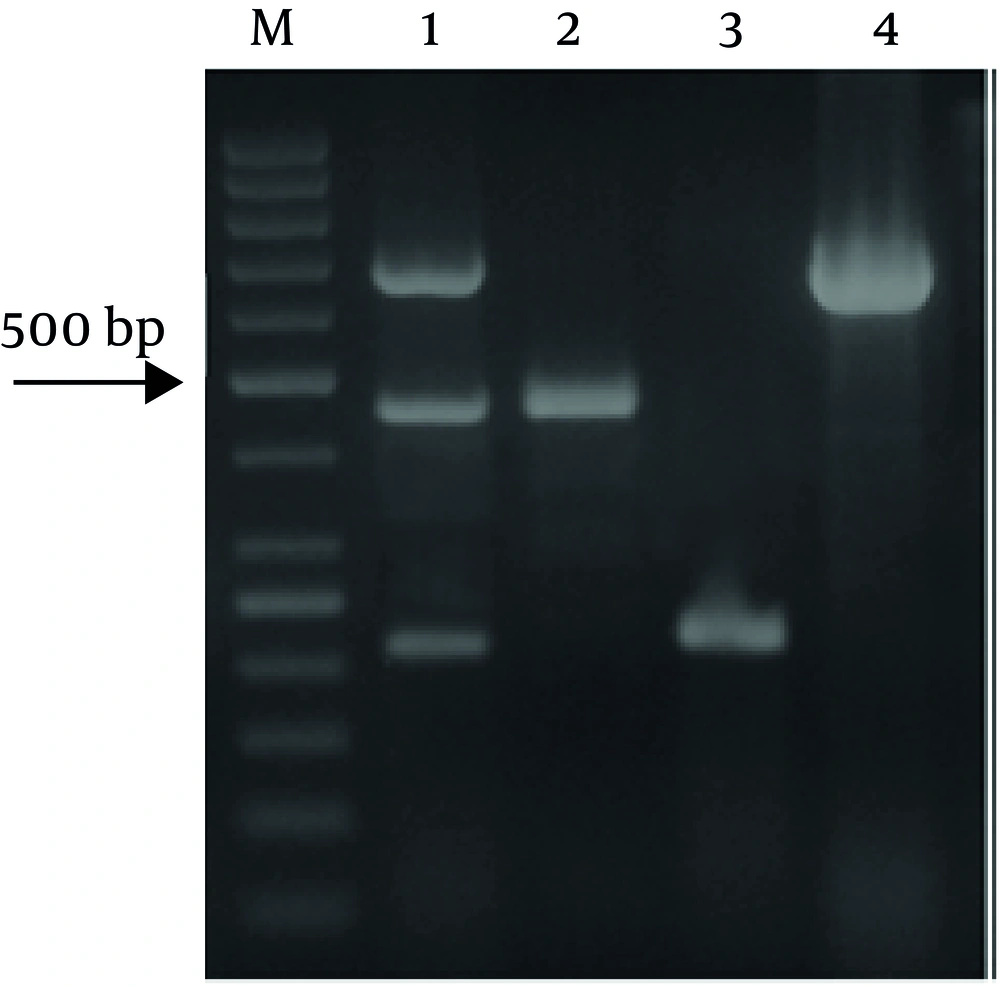
In the present study, the multiplex PCR-based method was used for detection of three food-borne pathogens after enrichment (Figure 1). The sensitivity of the multiplex PCR was assessed by artificially contaminated samples (Table 3). As shown, after enrichment, the method was able to detect the pathogens at the tested contamination levels in beef, buffalo and lamb samples. the prevalence of E. coli O157:H7, L. monocytogenes and Salmonella spp. in different kinds of meat were shown in Table 4 by using this method.
| Pathogen Used | Product | Level of Contamination(CFU/25 g) | PCR Resultsa |
|---|---|---|---|
| L. monocytogenes | Beef | 0 / 13 / 52 / 96 | − / + / + / + |
| Buffalo | 0 / 13 / 52 / 96 | − / + / + / + | |
| Lamb | 0 / 13 / 52 / 96 | − / + / + / + | |
| E. coli O157:H7 | Beef | 0 / 8 / 39 / 87 | − / + / + / + |
| Buffalo | 0 / 8 / 39 / 87 | − / + / + / + | |
| Lamb | 0 / 8 / 39 / 87 | − / (+) / + / + | |
| Salmonella spp. | Beef | 0 / 16 / 75 / 112 | − / + / + / + |
| Buffalo | 0 / 16 / 75 / 112 | − / + / + / + | |
| Lamb | 0 / 16 / 75 / 112 | − / (+) / + / + |
aadded after autoclaving and cooling to below 45 °C
| Types of Meat | Samples Tested, No. | Samples (%) | ||
|---|---|---|---|---|
| L. monocytogenes | E. coli O157:H7 | Salmonella spp. | ||
| Beef | 70 | 2 (2.8) | 2 (2.8) | 3 (4.3) |
| Buffalo | 70 | 2 (2.8) | 1 (1.4) | 2 (2.8) |
| Lamb | 70 | 3 (4.3) | 0 (0) | 5 (7.1) |
5. Discussion
Meat contaminations have been linked to consumer health problems, as reported by outbreaks and recalls from marketplaces associated with contaminated products. Measuring the microbial content of meat products is an important concern of the meat industry. Although classical microbiological methods offer reliable and standardized procedures for the detection of food-borne pathogens (e.g. ISO standards), they often including time consuming analyses that are not always compatible with the need for rapid results.
In this study, multiplex PCR as a rapid and cost-effective method, was used for the detection of pathogens in different kinds of meat. This assay was carried out according to Kim et al. procedure (5). They explained the sensitivity and specificity of this multiplex PCR assay as well as the validity of this assay for ham, milk and water samples. We assessed the sensitivity of this method by using artificially contaminated beef, buffalo and lamb samples. According to our results (Table 3), the sensitivity of this method along with its rapid and cost-effective results have make it a suitable method for the detection of these pathogens in meat samples. Therefore, the prevalence of E. coli O157:H7, L. monocytogenes and Salmonella spp. in beef, buffalo and lamb samples was assessed using this method.
As shown in Table 4, 2.8 % of the beef and 1.4 % of the buffalo samples contaminated with E. coli O157:H7. However, no contamination was found in the lamb samples. E. coli O157:H7 can colonize the intestinal tract of many livestock and during slaughtering may contaminate the carcass, work surfaces and material used for processing of meat products. With good hygienic practices during skinning and eviscerating, the rate of carcass contamination should be significantly below the carriage rate.
According to Breum and Boel, 3.2% of the Danish beef carcasses were contaminated with E. coli O157:H7 (18). However no contamination with E. coli O157:H7 was detected in 780 beef carcasses in Northern Ireland (13). In the United States, E. coli O157:H7 was detected in 1.6 and 2.9% of the pre evisceration and post intervention lamb carcasses, respectively, while in South Yorkshire, this pathogen was isolated from 1.4% of beef, and 0.7% of lamb carcasses (19, 20). According to Battisti et al., 2006, the overall prevalence of E. coli O157:H7 in slaughtered lambs in Rome was 0.2% (21).
L. monocytogenes can be found in a wide variety of raw and processed foods. Various meats and meat products such as beef, lamb, pork, and chicken, milk and dairy products, seafood and fish products have all been associated with Listeria contaminations (22). We have previously reported that 1.4% of raw/fresh fish and shrimp samples in this area contaminated with this pathogen (23).
In the present study, L. monocytogenes was detected in 2.8% of the beef and buffalo samples, and 4.3% of the lamb samples (Table 4). According to the previous reports from Iran, 2.6% of the beef and 6% of the lamb samples were contaminated with this pathogen (12 ). No contamination with L. monocytogenes was reported in 200 beef carcasses, in Northern Ireland (14 ). However, 0.42% of bison carcasses in USA were contaminated with this pathogen (24 ).
Salmonella spp. are among the most important food borne pathogens in the world. Poultry and poultry products are usually causing human salmonellosis outbreaks. According to our previous report (23), 2.9% of fish and 4.3% of shrimp samples in this area contaminated with Salmonella spp. In the present study, this pathogen was detected in 4.3% of the beef, 2.8% of the buffalo and 7.1% of the lamb samples. In the UK, Salmonella was detected in 1.7% of lamb, 1.1% of beef and 1.9% of pork samples (25). Other studies in the US and Italy also found the rate of Salmonella contamination to be higher in pork (3.3–9.9%) compared to beef products (1.0–1.9%) (26,27). The average Salmonella prevalences were 4.3 and 1.8% for pre evisceration and post intervention lamb carcasses, respectively (20).
This study shows that multiplex PCR is a very useful tool for the detection of food-borne pathogens, specifically L. monocytogenes, E. coli O157:H7 and Salmonella spp. The method represents a rapid and cost-effective way for detection of these pathogens in different kinds of meat.
When comparing our results to other authors, the discrepancies could be partly due to differences in sampling techniques and the detection methods. Besides, slaughter hygiene, cross contamination of the products at different stages throughout the food chain should be considered.
Overall, the results of the present study can provide a significant contribution to both regulatory agencies and the meat industries. Due to the potential hazard of these pathogenic bacteria, it is necessary to put more emphasis on meat hygiene. Therefore, the surveillance of potential contaminant bacteria in different kinds of meat is crucial to safeguard the public health.