1. Background
Aseptic meningitis is a common syndrome with acute onset of meningeal inflammation symptoms, fever, and pleocytosis of the cerebrospinal fluid (1, 2). Viral and bacterial meningitis cannot be differentiated properly. However examination of cerebrospinal fluid could result in excluding bacterial meningitis, and identification of the specific viral causes (1, 3, 4). Enteroviruses belong to the family of Picornaviridae, non-enveloped, single-stranded positive sense RNA viruses, which represent the major causes of aseptic meningitis worldwide (4, 5); although, most enteroviral infections are subclinical and self-limited severe illnesses such as meningitis, encephalitis, and paralysis, are resulted from the infection of central nervous system (CNS) (5-8).
2. Objectives
Traditional methods of detecting enteroviruses such as viral culture are time-consuming, labor-intensive procedures especially for CSF samples (1, 2, 4, 5, 7-9). Enterovirus molecular assays are the most sensitive and specific ones and the results will be available within several hours of specimen collection (10-12). Detection of the 5` non-coding region of the Enterovirus genome in CSF is the golden standard for the diagnosis of Enterovirus infection. Serotype identification should be conducted by sequence analysis of the amplified VP1 region as the major surface-neutralization epitopes in the Picornaviridae that correlates with the serotype classification (1, 2, 10). This study has been conducted to identify the prevalent serotypes of enteroviruses in young children with aseptic meningitis during 2009 - 2010 in Iran.
3. Patients and Methods
Cerebrospinal fluid specimens were collected from 118 children younger than 13 years of age with clinical suspicion of viral meningitis [fever, vomiting, headache, signs of meningeal inflammation, CSF WBC count > 5 × 106/mL, and a negative CSF culture for bacterial infection (Tuberclosis)]. They were negative for tumoral etiology as well. None of them were immunocompromised. All patients had been diagnosed by the specialists (Pediatrics and/or infectious disease specialist). Their specimens were transported on ice to the virology department of Pasteur Institute of Iran and stored at -80°C for further process. Complete clinical data were gathered from patients' medical records.
3.1. RNA Extraction
Genomic RNA was extracted from 200 µL of CSF samples and the vaccine vial of Oral Polio Vaccine (OPV) as a positive control, by using High-pure viral nucleic acid kit (Roche, Germany), according to the manufacturer’s instructions. To recover the extracted RNA, it was stored in nuclease-free water at -80°C.
3.2. Multiplex Real-Time RT-PCR
Extracted RNA was incubated at 75°C for 5 minutes and then was immediately cooled on ice for 3 - 5 minutes. cDNA synthesis was carried out in a 20 µL reaction volume containing 4 µL Revert Aid RT buffer (Fermentas Life Science, Lithuania ), 2 µL of 10 mM deoxynucleoside triphosphate mixture (dNTPs) (Fermentas Life Science, Lithuania), 0.5 µL of 40 U/µL RNase inhibitor protector (Fermentas Life Science, Lithuania), 1 µL of 200 U/µL RevertAid RT (Fermentas Life Science, Lithuania), 0.5 µL Random Hexamer primer, 8 µL, and 1 µL of the extracted RNA from CFS specimens and the positive control (OPV) respectively. The mixture was incubated at room temperature for 8 - 10 minutes and then 42°C for 60 minutes.
Afterwards, the tubes were inactivated at 95°C for 10 minutes. The Taq Man amplification was performed in a 25 µL reaction volume containing 12.5 µL 2x Premix ExTaq (TaKaRa, Japan), 1 µL of 10 pmol/µL primers and Probes (EV1; 5́-GGCCCTGAATGCGGCTAAT-3́, EV2; 5́-GGGATTGTCACCATAAGCAGCC-3́, (10), and 0.5 µL of 10 pmol/µL Taq Man probes; FAM 5́-AACCGACTACTTTGGGTGTCCGTGTTTC-3́ TAMRA, and 3 µL of the cDNA product. To detect any probable inhibitor and confirm the quality of extraction, 3 µL of an eGFP plasmid was added in each sample during the process of RNA extraction. The extracted eGFP plasmid was diagnosed by using specific primers (eGFP1; 5́-CTGCTGCCCGACAACCA-3́, eGFP2; 5́-ACCATGTGATCGCGCTTCTC-3́), and probe (HEX 5́-CCAGTCCGCCCTGAGCAAAGA-3́ BHQ) (11).
3.3. VP1 Amplification and Serotyping
Amplification of the enterovorises VP1 gene can be used to distinguish the Enterovirus serotypes. In this study the procedure for Nested-PCR amplification of the VP1 gene was optimized. cDNA was synthesized as described previously (12-14). The first PCR was carried out in a 25 μL reaction volume containing 10x PCR Buffer, 10 mM dNTP, 50 mM MgCl2, 5 U/μL Taq DNA polymerase (Cina Gen, Iran), 10 pmol of 224 primer: 5´-GCIATGYTIGGIACICAYRT-3´ and 222 primer: 5´-CICCIGGIGGIAYRWACAT-3´, and 5 μL of cDNA as template (12-14). This reaction was amplified in a Mastercycler Gradient (Eppendorf, Germany) by using the following conditions: initial denaturation at 95°C for 5 minutes, 45 cycles at 95°C for 50 seconds, 42°C for 50 seconds and 72°C for 1 minute and 10 seconds, plus a final extension step at 72°C for 10 minutes.
Electrophoresis of this first PCR product was run on 1.5% agarose gel composed of 0.5 μg/mL of ethidium bromide (Cina Gen, Iran), and the 850 base pair (bp) band was observed. Second round of the Nested PCR was performed using the following primers: AN 88 (5́-CCAGCACTGACAGCAGYNGARAYNGG-3́) and AN89 (5́-TACTGGACCACCTGGNGGNAYRWACAT-3́) (12-14), under the following conditions: initial denaturation at 95°C for 5 minutes, 40 cycles at 95°C for 45 seconds, 61°C for 45 seconds and 72°C for 45 seconds, plus a final extension step at 72°C for 7 minutes. The final 350 bp PCR product was analyzed by electrophoresis on 1.5 % agarose gel containing 0.5 μg/mL ethidium bromide (Cina Gen, Iran). The VP1 PCR products of each isolate were sent to Korea for sequencing. They were purified by using ABI purification kit and sequenced in both directions on an ABI Prism automated sequencer by using primers AN88 and AN89.
Enteroviruses serotypes were determined by pairwise comparison of the partial VP1 sequences to the available Enterovirus VP1 sequences in GenBank using FASTA program. Phylogenetic analysis was carried out to verify the typing results. The tree was constructed by neighbor-joining method using Mega 5 program. The robustness of phylogenies constructed with Neighbor was estimated by bootstrap analyses with 1000 pseudoreplicate data sets that support each node. References and GeneBank accession numbers for the sequences used in VP1 gene comparisons have been depicted in a phylogenic construct.
3.3.1. Nucleotide Sequence Accession Numbers
The sequences reported here were deposited in the GenBank sequence database under accession numbers as the following order: echo 30: JN092125. Two Polioviruses type 1: JN092124.1, and JN092123.1. Two echo 14: JN092122.1, and JN092121.1, and echo 5: N092120.1.
4. Results
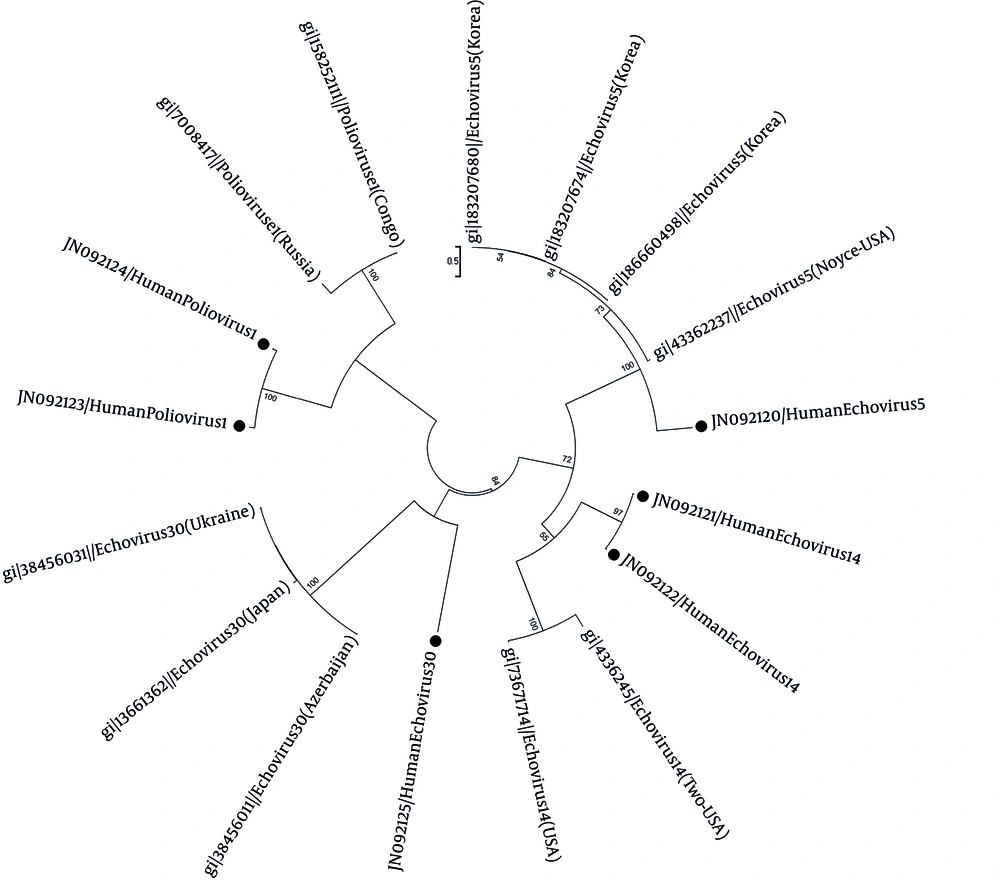
Sixty six patients were male (55.93%), and 52 were female (44.06%). Enteroviruses were detected in 12 (eight males, four females) out of 118 patients (10.16%). Enteroviral infection shows seasonal distribution. However, there was no significant seasonality in this study. The highest EN detection rate was in children under two years of age (18.75%), without any significant relationship between age and EN prevalence. Enterovirus serotyping was achieved only in six specimens that may be due to the insufficient CSF specimen volume or very low viral load in these specimens. The identified EN serotypes consist of one Echo 14 (8.3%), two polio enteroviruses type 1 (vaccine type) (16.6%), one Echo 30 (8.3%), and two Echo type 5 (16.6%). Phylogenic analysis showed similarities between the VP1 sequence of these isolates with the isolates from USA, Congo and Russia, Azerbaijan, Ukraine, Japan, and Korea (Figure 1).
The tree was elaborated by the neighbor-joining method using Mega 5. The bootstrap (1000 replicates) supporting each node is indicated. The scale bar corresponds to 0.5 substitutions/site. References and GeneBank accession numbers for the sequences used in VP1 gene comprised of; echovirus 5: 183207680 (Korea), 183207674 (Korea), 186660498 (Korea), 4336237 (USA). Echovirus 14: 4336245 (USA), 73671714 (USA). Echovirus 30: 38456011 (Azerbaijan), 13661392 (Japan), 38456031 (Ukraine). Poliovirus 1: 7008417 (Russia), 158252111 (Congo). Enteroviruses analyzed in this study are indicated by circle with their genotypes.
5. Discussion
Enteroviruses enter the human body usually through the oral-fecal route, but can also spread through the respiratory canals (3, 4). Enteroviruses are the major cause of aseptic meningitis, accounting for 85% to 95% of all cases in which a pathogen is identified (3-5, 9). The overwhelming meningitis cases are mostly caused by serotypes of coxsackievirus and echovirus. In Iran molecular epidemiologic data about the circulating enteroviruses and their role in viral meningitis is poor and results in unnecessary hospitalization of children and occurrence of the secondary infections (as the nosocomial infections).
During 2007 - 2008 enteroviruses were detected in 30 out of 65 children with viral meningitis in south of Iran (Shiraz city in Fars province) followed by other viruses (46.2%) (15). In another study from south of Iran (Ahvaz city in Khuzestan province) enteroviruses were detected in 52.63% of CSF specimens (16). There are also other reports from Ahvaz about the frequency of enteroviruses in one of the hospitals in Ahvaz and the frequency of Echo 30 in enteroviral meningitis patients (17, 18). Another study showed that the frequency of enteroviruses in viral meningitis in Tehran is similar to other countries (1, 2, 4-7).
Enteroviruses circulate with different patterns in different years and regions even inside a country, therefore, it is anticipated that different epidemiological reports are available (3, 5, 8). Since differentiation of enterovirus serotypes is based on neutralization and the VP1 sequence correlates with neutralization type, molecular diagnoses of the VP1 region correlates with the serotype and are used for differentiation in the clinical laboratories (4, 10-12). Many studies have shown that enterovirus type B is the major cause of viral meningitis (1, 2, 4, 5, 7, 9). In an outbreak of viral meningitis in Greece, poliovirus type 1 (vaccine type),
Echoviruses types 14 and 30 were detected in 2001 and 2006, respectively (19). In 2007, echovirus 4 was the predominant serotype in Greece and other serotypes such as echovirus 6, 9, 14, 25, Coxsackie A6, A15, A24 and Coxsackie B1 were identified as the circulating serotypes (19). Echo virus 30 was the most frequent serotype in Spain in 2006 (4).
The serotypes of detected enteroviruses in this study were identified using genetic analysis of the VP1 region to determine the circulation pattern and the phylogenetic relation between isolated enteroviruses from distant geographical areas, and the VP1 region was also localized within the B cluster (coxsackievirus group B- or echovirus-like genotype).
Phylogenic analyses of these EN serotypes show similarities between the two isolated polioviruses type 1 in this study and the isolates from Congo and Russia. The two isolated echo 14 strains are similar to the echovirus 14 strains isolated from USA. There is also a relation between the echo 30 isolated in this study and isolates from Japan, Azerbaijan and Ukraine. The echo 5 isolate is similar to the isolates from Korea and USA. Although, the direct genotype of enteroviruses of CSF specimens was conducted for the first time in Iran in this study and the results of genotype assessments are not comparable with previous data from Iran, but these findings provide valuable information to improve our knowledge about enteroviruses epidemiology, their possible relationship with neurological diseases and the need for further studies in Iran.