1. Background
Cutaneous Leishmaniasis (CL) continues to be an increasing public health problem in Iran (1). CL is endemic in half of the 30 Iranian provinces (2). Recently, several new foci have been reported, indicating the potential spread of the disease in the country (3). Both epidemiological forms of CL are present in Iran; Zoonotic Cutaneous Leishmaniasis (ZCL) caused by Leishmania major and Anthroponotic Cutaneous Leishmaniasis (ACL) due to L. tropica (4). Essentially, identification of Leishmania parasites is necessary for epidemiological objectives such as documenting the distribution of prevalent species and designing appropriate control measures, and it is also important for treatment modality (5-7). Leishmania parasites have similar morphology and sometimes cause similar clinical manifestations; therefore, differentiation among species requires molecular techniques such as Polymerase Chain Reaction (PCR) (8).
So far, some different molecular techniques such as RFLP (9-11), RAPD (5, 12, 13), Real-time PCR (14, 15) and Nested PCR (1, 16-19) have been performed regarding the identification of Leishmania in human specimen by species level. A Nested PCR based method that permits both very sensitive detection and high-resolution identification of Leishmania parasites directly from clinical samples is presented here (20). This is the first study of molecular identification of Leishmania species in Poledokhtar district, also Lorestan province.
2. Objectives
This cross-sectional study was conducted to identify the Leishmania spp. isolated from CL patients who referred to Poledokhtar health centre, using Nested PCR method.
3. Materials and Methods
3.1. Sampling and Study Area
This descriptive study was performed on 52 patients infected by Leishmania and referred to Poledokhtar health centre laboratory in 2008-2011. The slide smears of lesions patients suspected to CL were prepared using Giemsa staining. Then the leishman bodies were observed using common microscopic technique and they were subjected to molecular technique to be identified. The Poledokhtar district (33° 9′ 13″ N, 47° 42′ 49″ E at an altitude of about 660 m above sea level) lies in the south of Lorestan province. The weather is hot in the summer and moderate in the winter.
3.2. DNA Extraction
The smear scrapings were added to 150 µl lysis buffer (50mm Tris-Hcl pH 7.6, 1mm EDTA pH 8.0, 1% Tween 20, 8.5 µl proteinase K solution 19 mg/mL) and incubated for 2 h at 55°C. Phenol-chloroform- isoamyl alcohol extraction method was used to extract DNA. The DNA samples were dissolved in 50μl deionized distilled water and stored at 4°C (21).
3.3. Nested-PCR
Special primers related to variable regions of kDNA were used in a Nested-PCR technique. The external primers CSB1XR (CGA GTA GCA GAA ACT CCC GTT GA) and CSB2XF (ATT TTT CGC GAT TTT CGC AGA ACG) in the first round and internal primers LiR (TCG CAG AAC GCC CCT) and 13Z (ACT GGG GGT TGG TGT AAA ATA G) in the second round were applied.DNA was amplified using programmable Thermocycler (Thecne Cambridge, UK) under the following conditions: 5 min at 94°C followed by 35 cycles of 30 sec at 94°C, 60 sec at 55°C, 90 sec at 72°C and a final elongation at 72°C for 10 min.
3.4. Agarose gel Electrophoresis
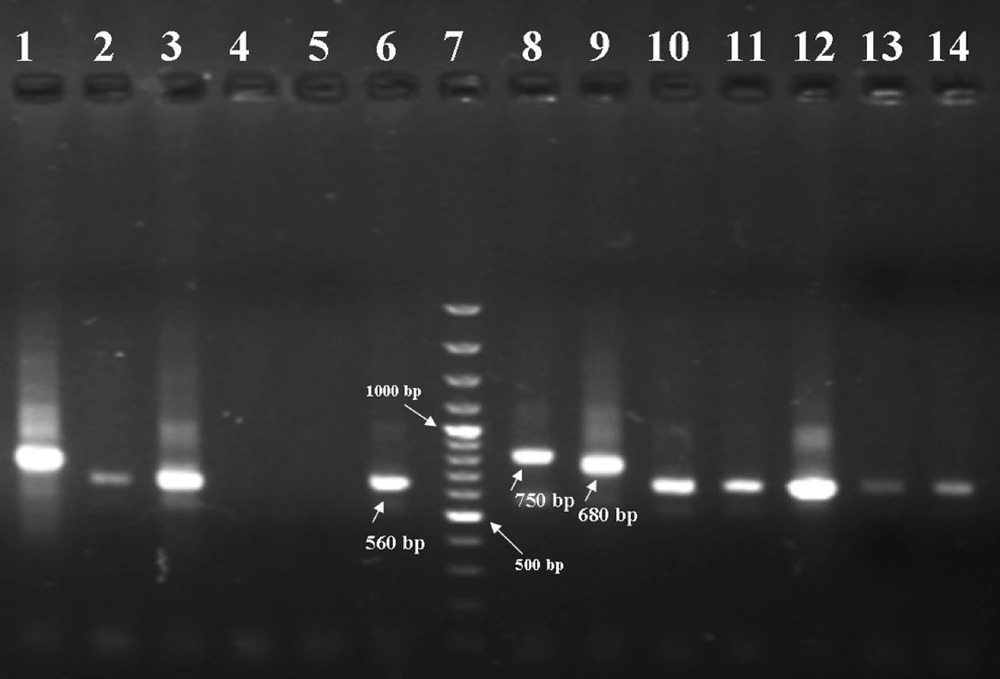
The PCR products were visualized by 1.5% agarose gel electrophoresis (Uvitech, Cambridge UK), using a 100 bp DNA ladder marker at 260 nm wavelength. A negative control and three positive controls including L. major (MHOM/IR/54/LV39), L. tropica (MHOM/IR/89/ARD2) and L. infantum (MCAN/IR/96/Lon49) were used in each round of PCR and electrophoresis. L. tropica, L. major and L. infantum provided fragments of 750 bp, 560 bp and 680 bp respectively.
4. Results
From 52 confirmed parasitological cases, 59.62% were male and 40.38% were female (Table 1). The results of the second-round PCR showed that L. tropica generated a 750bp fragment whereas L. major generated a 560bp (Figure 1). Comparison of the pattern of electrophoretic profile of the studied isolates with the electrophoretic pattern of reference strains helped to identify that from 52 isolates, the prevalence of L. major was 50 (96.15%) and that of L. tropica was 2 (3.85%) among the cases. L . major isolates were from some parts of Poledokhtar. The more detailed information related to the different parts of the studied area is presented in the Table 2.
| Clinical form of the lesion | Male | Female | Total, No. (%) |
|---|---|---|---|
| Dry | 4 | 4 | 8 (15.38) |
| Moist | 27 | 17 | 44 (84.62) |
| Total | 31 | 21 | 52 (100) |
L. tropica isolated from the patient (lane 1), L. amajor isolated from the patients (lanes 2, 3, 10, 11, 12, 13 and 14), Negative controls (lanes 4 and 5), reference strain of L. major (560 bp) (lane 6), marker (100-bp ladder) (lane 7), reference strain of L. tropica (750 bp) (lane 8), reference strain of L. infantum (680 bp) (lane 9).
| Part | Cases | L. major, No. (%) | L. tropica No. (%) |
|---|---|---|---|
| City Centre | 6 | 6 (100) | - |
| Mamoolan | 4 | 3 (75) | 1 (25) |
| Afrineh | 6 | 5 (83.33) | 1 (16.67) |
| Malavi | 12 | 12 (100) | - |
| Jaidar | 11 | 11 (100) | - |
| Jelogir | 4 | 4 (100) | - |
| Western Miankouh | 8 | 8 (100) | - |
| Eastern Miankouh | 1 | 1 (100) | - |
| Total | 52 | 50 (96.15) | 2 (3.85) |
5. Discussion
Accurate identification of the Leishmania species seems to be necessary for a variety of clinical and epidemiological reasons to decide on distinct treatment regimens and also design appropriate control programmers (22). DNA-based techniques have been commonly used as potential tools for this purpose (23). The current study showed that ZCL was prevalent in Poledokhtar district. It is recommended that more studies should be carried out to identify vectors and reservoirs of L. Major in the Poledokhtar, and according to the type of parasite and its reservoirs CL control programs should be applied in this area. The obtained results of the current investigation are in consistence with the studies of Maraghi et al. (16), Razmjou et al. (1), Alimoradi et al. (12), Azizi et al. (17), Khosravi et al. (14), Rahbarian et al. (24), Mohamadi Azni et al. (18), Maraghi et al. (25), and Ghasemian et al. (26) that they had reported the frequency of L. major from 86-100%.
However, the current study results were different from Hajjaran et al. (5), Sharifi et al. (19) and Mohajeri et al. (13), The results of the current study indicated that most of the recognized cases of CL in Poledokhtar were due to L. major. In conclusion, characteristics of the collected Leishmania isolates showed that L. major is predominant agents of CL, like other western and southeastern regions. Based on the studies conducted in the southwest and west of the Country in Khuzestan, Kermanshah and Ilam Provinces, L. major isolates were recovered and identified from CL patients (12, 16, 27) and L. major recovered from wild rodents like Tateraindica, and Merioneslibycus as the principal reservoirs host in this area (28). Further studies on reservoir and vector of ZCL are necessary to better clarify the epidemiological aspect of leishmaniasis in Poledokhtar district.