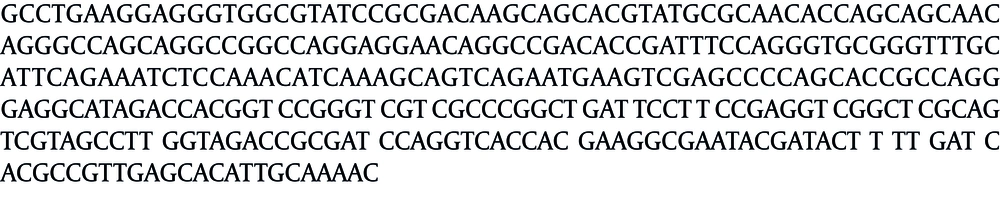1. Background
Extended-spectrum β-lactamases (ESBLs) are a group of enzymes that mediate resistance to extended-spectrum cephalosporins, such as ceftriaxone, cefotaxime, ceftazidime, and the monobactam aztreonam. Clavulanic acid-inhibited extended-spectrum β-lactamases (ESBLs) have been described primarily in Enterobacteriaceae then in Pseudomonas aeruginosa (1, 2). Beta lactamase TEM (Temoneria) and SHV (refering to sulfhydral variable) have been known as parental ESBL enzymes. Other types of ESBL enzymes such as PER (Pseudomonas extended resistance), GES (Guiana Extended-Spectrum), certain OXA type enzymes (preferentially hydrolysis oxacillin and cloxacillin), have also been explained in some of countries (1).
GES, the new class of ESBLs, was reported in 2000 in a clinical isolate of Klebsiella pneumoniae from French Guiana (2). Identification of GES-2 is an interesting research progress on ESBLs in P. aeruginosa (3). The GES-2 β-lactamase is a class A carbapenemase, the emergence of which is an alarming advance in clinically considerable bacterial pathogens as the enzyme confers resistance to carbapenem antibiotics (4). The integron genetic framework of GES-2 is the essential factor that emerges resistance to broad-spectrum β-lactam antibiotics and other dissimilar classes of antimicrobials (3).
Outcome of introducing such characteristics brings out a huge challenge around effective treatment and control of these isolates on a health system (5). The blaGES-2 is located on a gene cassette in class 1 integron. For the first time, GES-2 was reported in MDR clinical isolates of P. aeruginosa, originating from a university hospital in South Africa (6). Poirel et al. reported that GES-2 was associated with an outbreak occurring in the same hospital in 2000 (7). Rapid recognition of resistant strains and detection of resistance genes is necessary to succeed in antimicrobial therapy and infection control policy. Although, epidemiological data indicate that ESBL-producing P. aeruginosa has sparse prevalence in Iran.
2. Objectives
This study was performed to investigate the presence of β-lactamase blaGES-2 gene in ESBL-producing isolated P. aeruginosa from a tertiary care teaching hospital in Kashan, Iran, DNA sequencing for selected PCR products of the blaGES-2 gene, sequence comparing and aligning with reference sequence and evaluate their resistance to eight commonly-used antibiotics.
3. Materials and Methods
A total of 100 clinical and environmental isolates of P. aeruginosa were non-repetitively and consecutively obtained from clinical specimens and wet environment of Beheshti hospital in Kashan, Iran, from February 2010 through September 2011. Species identifications were done by standard methods. Eight ESBL-producing isolates of P. aeruginosa were detected. The strains were isolated from blood [1], trachea [2], urine [2], gastrointestinal fluid [1], bronchoscopy fluid samples [1] and wet environment of the hospital [1].
Antimicrobial susceptibility testing was performed on all eight isolates according to the standard method established by Clinical and Laboratory Standards Institute (CLSI) (8). Imipenem (10 μg), ciprofloxacin (5 μg), ceftazidime (30 μg), ceftriaxone (30 μg), cefotaxime (30μg), aztreonam (30 μg), piperacillin (100 μg) and gentamicin (10 μg) [Mast Group Ltd., Merseyside, UK] were determined by disk diffusion method. Multi-drug resistance was defined as resistance to three or more classes of antibiotics. The quality control of antibiotic susceptibility was determined by P. aeruginosa ATCC 27853. Double disk synergy test was used for the assessment of ESBLs production.
Detection of β-lactamase blaGES-2 gene was performed under standard PCR conditions using the published set of primers (Table 1). The primers used for these tests were obtained from Cinagene Company, Iran. To perform PCR, a 1:10 dilution of a 24-hour culture was boiled for 10 minutes. Then amplification was performed with 1:10 of this dilution as the DNA template. PCR settings involved 30 cycles of amplification under the following conditions: Denaturation at 95°C for 30 seconds, annealing at 50°C for 60 seconds and then at 72°C for 60 seconds. Then, cycling was followed by a final extension at 72°C for 5 minutes. The PCR products were analyzed by electrophoresis on a 1.5% agarose gel. After electrophoresis, the gel was stained using ethidium bromide and photographed under a UV trans-illuminator (Ingenius, Syngene).
A 100-base pair (bp) DNA (Bioneer, Korea) ladder was used as the molecular size marker. DNA sequencing for selected PCR products of the blaGES-2 gene was performed for identification of detected bla gene using primers as shown in Table 1. For this matter, the PCR products of the above-mentioned gene were subjected to direct sequencing of both strands using an automated sequencer (ABI system, 3730XL) of Macrogen Company, Korea. The nucleotide sequences were analyzed by Chromas LITE software, version 2.01. Sequences were compared and aligned with the reference sequence using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and CLUSTAL W (http://www.ebi.ac.uk/clustalw).
| Gene Type | Primer | Sequences (5´ to 3´) | Product Size, bp | Reference |
|---|---|---|---|---|
| GES-2 | GES-2 | GTTTTGCAATGTGCTCAACG | 371 | (9) |
| AGES-2 B | TGCCATAGCAATAGGCGTAG |
The nucleotide sequences of blaGES-2 gene and the partial gene encoding ABC transporter permease (G-1) were deposited in Gene Bank through DNA Data Bank of Japan (DDBJ), and the accession numbers assigned were AF326355.1 and AB591379.1, respectively.
4. Results
The prevalence of ESBLs in P. aeruginosa isolates was 8%. The resistance pattern of eight ESBL-producing P. aeruginosa isolates in our study showed an extra high resistance to piperacillin (75%) and all extended-spectrum cephalosporins (62.5%). The isolated ESBL-producing P. aeruginosa samples were more sensitive to imipenem, gentamicin and ciprofloxacin compared to other tested antibiotics (Table 2). Fifty percent of the isolates, (four out of eight) were resistant to at least three classes of antibiotics and classified as multi-drug resistant (MDR). By using the blaGES-2primer pair, visible bands were observed near the location of the expected size in eight isolates. In order to increase the accuracy of the results, sequencing of both forward and reverse strands of the blaGES-2 gene were carried out twice (Figure 1). Sequence alignment with the reference sequence of blaGES-2 (Accession no. AF326355.1) by BLASTn search and CLUSTAL W revealed no similarity and BLASTn search identified them as the ATP-binding cassette (ABC) transporter permease, partial cds, clone G-1 P. aeruginosa (Accession no. AB591379.1), with 84% coverage and 100% maximum identity.
| Antibiotics | Sensitive, No. (%) | Intermediate, No. (%) | Resistant, No. (%) |
|---|---|---|---|
| Piperacillin (100 g) | 2 (25) | 0 (0) | 6 (75) |
| Imipenem (10 g) | 4 (50) | 0 (0) | 4 (50) |
| Gentamicin (10 g) | 4 (50) | 0 (0) | 4 (50) |
| Cefotaxime (30 g) | 1 (12.5) | 2 (25) | 5 (62.5) |
| Ceftriaxone (30 g) | 2 (25) | 1 (12.5) | 5 (62.5) |
| Ceftazidime (30 g) | 2 (25) | 1 (12.5) | 5 (62.5) |
| Aztreonam (30 g) | 3 (37.5) | 1 (12.5) | 4 (50) |
| Ciprofloxacin (5 g) | 4 (50) | 3 (37.5) | 1 (12.5) |
5. Discussion
In our study, blaGES-2 gene was detected in all ESBL-producing isolates. GES β-lactamase genes which are placed on the broad-host-range conjugative plasmids, are the gene cassettes elements of class 1 integrons found in gram-negative isolates and have been spread worldwide (4). GES-2 β-lactamase is the fourth example of a non-TEM/ non-SHV-type ESBL in P. aeruginosa after PER-1, VEB-1 (Vietnamase extended spectrum) and OXA-18 (6). GES-type β-lactamase consists of 15 members (GES-1 to GES-15), isolated from various Gram-negative bacteria in Europe, Asia, Africa, and America (4). Although some GES β-lactamases produce antibiotic resistance profiles like those of classical extended spectrum enzymes, some GES variants (GES-2, 4, 5, and 6) are conferred with reduced susceptibility to imipenem (4).
GES β-lactamases are found in French Guiana, Greece, and South Africa (10). blaGES-1 has also been found in a P. aeruginosa strain isolated in a French medical center and from a 63-year-old female who had a hysterectomy (Sao Paulo, Brazil) (9). GES-2 was identified from a P. aeruginosa isolate in South Africa and later from eight more strains involved in the outbreak. GES-2 β-lactamase may supply partly to the decreased susceptibility of P. aeruginosa to imipenem (6, 7, 11). GES-2 β-lactamase belongs to the class A carbapenemase, the emergence of which is an alarm in P. aeruginosa as the enzyme confers resistance to carbapenem antibiotics. Tazobactam is a clinically used inhibitor of class A β-lactamases, which inhibits the GES-2 enzyme efficiently, retrieving the susceptibility to β-lactam antibiotics.
In our study, BLASTn search showed that the blaGES-2 gene sequencing matched as the ATP-binding cassette (ABC) transporter permease, partial cds, clone G-1 P. aeruginosa (Accession no. AB591379.1). Same results were obtained by Hirakawa et al. (12). Resistance in P. aeruginosa may be mediated via several distinct mechanisms including the production of β- lactamases, efflux pumps, and target-site or outer membrane modifications (13). Efflux pump systems have been revealed as the extremely important causes of multi-drug resistance in P. aeruginosa. ABC transporters are the major efflux pump protein families that mediate resistance to antibiotics and are believed to play a crucial role in the development of MDR (14, 15). In general, ABC transporters have been found in both prokaryotic and eukaryotic systems and are responsible for the import and export of various proteins, peptides, polysaccharides, and drugs that utilize ATP hydrolysis to drive the export of substrates (15).
Detection of the ESBL GES-2 from P. aeruginosa in seven hospitalized patients and one environmental sample clarified that these isolates had been established in our teaching hospital. GES-2-producing P. aeruginosa tends to colonize and mostly infects debilitated patients, considerably increasing both their lengths of hospitalization and costs of treatment. The integron genetic structures that support GES-2, present not only resistance to broad-spectrum β-lactam antibiotics but also to dissimilar classes of antimicrobials, making these isolates very difficult to treat and control successfully. In our study, prevalence of MDR P. aeruginosa isolates, was 50% which was lower than the value reported in the study conducted by Aggarwal et al. (100%) (16).
Carbapenems were subsequently introduced into the clinic as the last resorts of antibiotics due to their high potency and exceptional broad spectrum of antimicrobial activity that includes both gram-negative and gram-positive aerobic and anaerobic bacteria. New types of class A β-lactamases, the carbapenemases, are capable of causing resistance to a wide variety of β-lactam antibiotics, including carbapenems (4). In this study, notable resistance (50%) of P. aeruginosa was observed against imipenem, while other studies reported that more than 80% of ESBL producing isolates were sensitive to imipenem (17, 18). Our study reports the presence of blaGES-2 in P. aeruginosa for the first time in Iran. ESBLs are uncommon but significant problems in our hospital. GES-2 was found in all of ESBL-positive isolates, of which 50% appeared to be MDR.