1. Background
Acinetobacter baumannii is a problematic nosocomial pathogen, especially in patients admitted to intensive care units (ICUs), those requiring mechanical ventilation, and patients with wound or burn injuries. This microorganism causes life threatening infections such as bacteremia, pneumonia, meningitis, urinary tract and wound infections (1). A. baumannii possess a remarkable ability to acquire plasmids, transposons, or integrons that carry clusters of resistant genes, and this ability leads to multi drug resistance (MDR). Control of MDR in A. baumannii is a medical concern, because of the limited therapeutic choices available. It should be noted that increasing resistance to carbapenems has been observed worldwide in the past decade.
Carbapenemase production is the most described mechanism of resistance to carbapenems (2). Carbapenem resistance in A. baumannii is mediated by the acquisition of a class B or a class D β-lactamase such as oxacillinase (3). Since the first description of a carbapenem-hydrolyzing oxacillinase, in 1993, several oxacillinases with a carbapenem-hydrolyzing activity have been reported (4). Nowadays, OXA-type carbapenemases have been divided into eight subgroups which four of them have been identified in A. baumannii: OXA-23-like consist of (OXA-23, OXA-27 and OXA-49); OXA-24-like (OXA-24, OXA-25, OXA-26, OXA-40 and OXA-72); OXA-58; and OXA-51-like. The last group is a family of chromosomal enzymes typically found in A. baumannii. Acquired bla oxa-23 gene is located in transposon, mainly, Tn2006 (ISAba1 linked) and Tn2007 (ISAba4 linked). The strains which could produce Oxa-23, have been reported as sources of nosocomial outbreaks worldwide (2) In order to better understand and control multidrug-resistant A. baumannii, understanding the molecular basis of the infection is necessary.
2. Objectives
The current study described antimicrobial susceptibilities and conducted a multiplex PCR assay to detect alleles encoding oxacillinases. The study determines the prevalence of oxacillinase genes in clinical isolates of A. baumannii from some Tehran hospitals.
3. Materials and Methods
3.1. Bacterial Isolates
Acinetobacter spp. (n = 131) were isolated from L and M, hospitals in Tehran, during 2010–2011. These bacteria were originally isolated from aspirated sputum, trachea, burn, wound and urinary tract infections. The clinical Acinetobacter isolates, were primarily identified by Gram staining as Gram negative coccobacillary rods that may initially appear in direct smears, as Gram positive cocci, non motile on S.I.M medium, oxidize negative and lack of lactose fermentation (5).
3.2. DNA Extraction
Genomic DNA was extracted by boiling method. Briefly, five to six colonies were suspended in 250 µl sterile ultrapure water and boiled for 10 minutes. The samples were cooled on ice (10 minutes) and centrifuged at 14000 rpm at room temperature.The supernatant was transferred to a new tube and kept at4°c for further analysis (6).
3.3. Detection of blaOXA-51-Like Gene to Identify A. baumannii Species
All isolates were subjected to the PCR to detect blaoxa-51-like gene which is unique to A. baumannii species (7, 8).
3.4. MDR Definition
Multidrug resistance was defined in this analysis as resistance to three or more representatives of the following classes of antibiotics: quinolones (ciprofloxacin), extended-spectrum cephalosporins (ceftazidime), aminoglycosides (amikacin, gentamicin), and carbapenems (imipenem, meropenem) (9).
3.5. Susceptibility Testing
Susceptibility to conventional antibiotics was performed by the disk diffusion method as recommended by the (CLSI). Colistin (10 µg), Imipenem (10 µg), Meropenem (10 µg), Gentamicin (10 µg), Ciprofloxacin (5 µg), Amikacin (30 µg), Cotrimoxazole (25 µg), Cefepime (30 µg), Cefotaxime (30 µg), Aztreonam(30 µg), Ceftazidime (30 µg), and Polymyxin B (300 U) were obtained from Mast company (Pharmaceutical Inc. UK). Quality control was performed by testing the susceptibility of Escherichia coli ATCC 25922 (10).
3.6. Detection of Carbapenem-Resistant Genes
A multiplex polymerase chain reaction (PCR) assay was performed to detect the carbapenem-resistant genes in the A. baumannii isolates according to the method described by Woodford et al. ( 11 , 12 ). Primers were designed to amplify fragments of bla OXA-23, bla oxa-24,bla 0xa-58, carbapenemase genes. The amplification conditions were: initial denaturation at 94°C for 5 minutes, 30 cycles of 94°C for 25 seconds, 52°C for 40 seconds, 72°C for 50 seconds, and a final elongation at 72°C for 6 minutes (Table 1).
| Primers | Primer Sequence (5’-3’) | Product Size (Base pair) |
|---|---|---|
| OXA-51-like | TAATGCTTTGATCGGCCTTGTGGATTGCACTTCATCTTGG | 353 bp |
| OXA-23-like | GATCGGATTGGAGAACCAGAATTTCTGACCGCATTTCCAT | 501 bp |
| OXA-24-like | GGTTAGTTGGCCCCCTTAAAAGTTGAGCGAAAAGGGGATT | 246 bp |
| OXA-58-like | AAGTATTGGGGCTTGTGCTGCCCCTCTGCGCTCTACATAC | 599 bp |
Multiplex PCR Primers to Detect Genes Encoding oxa Carbapenemase
4. Results
4.1. Strain Identification
131 isolates of Acinetobacter, were identified by conventional identification methods. 91 (69.46%) isolates were obtained in the intensive care unit, 21 (16.03%) isolates in the internal wards, 10 (7.63%) isolates in the Surgical ward, 5 (3.8%) isolates in the emergency, 2 (1.52%) isolates in ENT, and 2 (1.52%) isolates, obtained in pediatrics. 7 (5.34%) isolates were obtained from sputum. 16 (12.21%) from burn, 65 (49.61%) from pleural effusion, and 21 (16.03%) isolates were obtained from wounds. In addition to them, there were isolates from urine, ear, and other samples. The blaOXA-51-like gene was amplified from genomic DNA to detect A.baumannii. 123 isolates (93.89%) that gave a band for bla OXA-51-like gene, were identified as A. baumannii and 84 (68.29%) of these isolates were found in ICU.
4.2. Detection of Carbapenem-Resistant Genes
Co-existence of different blaoxa genes among 123 isolates of A. baumannii and carbapeneme resistance of each isolate are indicated in Table 2. As observed in Table 2, among the four isolates which contain only blaOXA-51-like gene, no other oxacillinase genes, one isolate was carbapenem resistant. Along with other oxacillinase genes, increase in carbapenem resistance is observed. About bla OXA-23 gene, prevalence of gene and resistance rate is higher. None of the isolates carrying bla Oxa-58 associated with bla Oxa-23 or bla Oxa-24 genes. Table 3 shows the characteristics of isolates which possessed more than one of the carbapenemase genes. All of these isolates contain blaOXA-51-like gene, because this gene is intrinsic to all of A. baumannii isolates. These isolates have been isolated from two hospitals in Tehran named L and M. These isolates are resistant to three or more classes of antibiotics. According to the definition, it can be termed as MDR (9).
| Blaoxa genes | No. (%) | Imipenem Resistance only, No. (%) | Meropenem Resistance only, No. (%) | Resistance to Both Imipenem and Meropenem, No. (%) | Total, Resistance to Each of Carbapenem and to Both of Carbapenems, No. (%) |
|---|---|---|---|---|---|
| Oxa-51 only | 4 (3.25%) | 0 (0.0%) | 0 (0.0%) | 1 (25%) | 1 (25%) |
| Oxa51 and Oxa-23 only | 100 (81.3%) | 2 (2%) | 31 (31%) | 56 (56%) | 89 (89%) |
| Oxa-51 and Oxa-24 only | 10 (8.13%) | 0 (0.0%) | 5 (50%) | 3 (30%) | 8 (80%) |
| Oxa-51 and Oxa-58 only | 1 (0.81%) | 0 (0.0%) | 1 (100%) | 0 (0.0%) | 1 (100%) |
| Oxa-51 and oxa-23 and oxa-24 | 7 (5.69%) | 0 (0.0%) | 2 (28.57%) | 3 (42.8%) | 5 (71.42%) |
Distribution of Different blaoxa Genes Among Clinical Isolates of Carbapenem Resistance A. baumannii
| Isolated Name | Hospital | Ward | Specimen | Antimicrobial Resistance | Oxa Genes |
|---|---|---|---|---|---|
| 30T | L | medical | trachea | AN a, GM a, SXT a, CP a, CAZ a, CPM a, CTX a, ATM a | oxa-23, oxa-4, oxa-51 |
| 47T | L | medical | sputum | MEN a, SXT, CP, CAZ, CPM, CTX, ATM | oxa-24, oxa-23, oxa-51 |
| 76T | M | I.C.U | ear | AN, GM, MEN, SXT, CP, CAZ, CPM, CTX, ATM | oxa-24, oxa-23, 0xa-51 |
| 77T | M | I.C.U | trachea | AN, GM, IPM a, MEN, SXT, CP, CAZ, CPM, CTX, ATM | oxa-23, oxa-24, oxa-51 |
| 114T | M | I.C.U | burn | GM, SXT, CP, CAZ, CPM, CTX | oxa-23, oxa-24, oxa-51 |
| 129T | L | I.C.U | trachea | GM, IPM, MEN, SXT, CP, CAZ, CPM, CTX, ATM, | oxa-23, oxa-24, oxa-51 |
| 131T | L | I.C.U | catheter | AN, GM, IPM, MEN, SXT, CP, CAZ, CPM, CTX, ATM | oxa-23, oxa-24, oxa-51 |
Characteristics of Isolates Which Possessed More Than one of the Carbapenemase Genes
4.3. Antimicrobial Susceptibilities
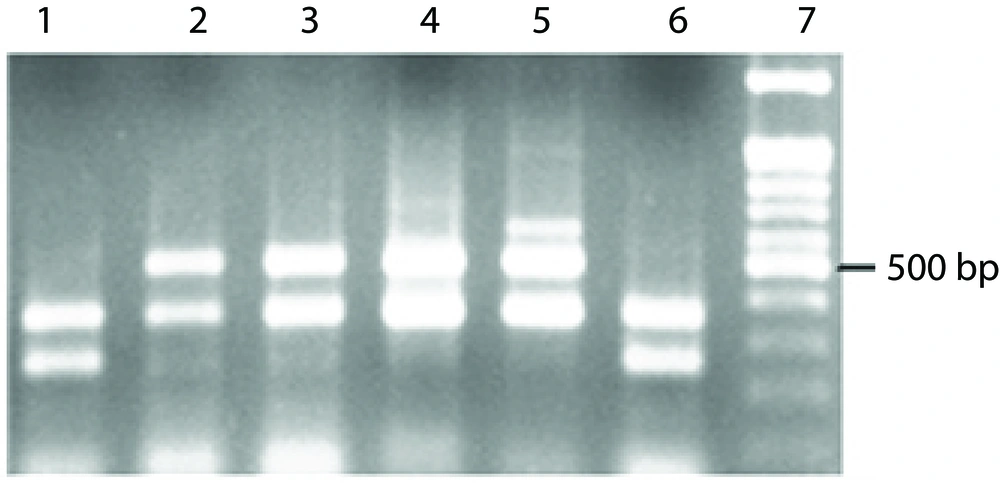
Isolates of A. baumannii showed, 118 (95.93%) to ciprofloxacin 95 (77.23%) to gentamicin, 100 (81.3%) to cefepime, 100 (97.56%) to aztreonam, 117 (95.12%) to ceftazidime 109 (88.61%) resistance to trimeto prime sulphametoxazole, 67 (54.47%) to amikacin, 83 (67.47%) to imipenem, and 104 (84.55%) to meropenem. All isolates were susceptible to colistin and Polymyxin B.43, of 123 A. baumannii isolates (34.95%) were MDR. These isolates were resistant to amikacin, ciprofloxacin, imipenem, meropenem, ceftazidim. 37 (86.04%) isolates of MDR A. baumannii possessed only bla oxa-23-like genes, and 2 (4.65%) possessed both oxa-24/oxa-23-like genes. One (2.35%) possessed only bla oxa-51-likegene (Figure 1).
5. Discussion
A . baumannii is considered as one of the most important nosocomial pathogens. The occurrence of MDR and pan drug-resistant A. baumannii is a growing concern. The current study indicated that, 43 of 123 A. baumannii isolates (34.95%) were MDR. These isolates were resistant to amikacin, ciprofloxine, imipenem, meropenem, ceftazidim. As Table 3 shows, 5 of 7 (71.43%) isolates which carried more than two oxacillinase genes, and were MDR, had been isolated from intensive care units. Resistance rates can differ according to the country, hospital under review, and depend on biological, epidemiological or methodical factors (13). In 1999, 15 hospitals in Brooklyn, reported high rates of MDR A. baumannii infection, twelve percent of the strains were resistant to all commonly used drugs (14 ). During the years 2003 to 2004, of A. baumannii isolates, 76% were multi-drug resistant (MDR); almost half of them being resistant to every tested antimicrobial except, imipenem (15).
In Washington DC in 2006, 89% of Acinetobacter isolates, were resistant to at least 3 drugs, meeting the criteria for multidrug resistance (16). The high rate of MDR isolates in studies referred to here between 2003 to 2006, compared to the MDR rate obtained in the current study is due to the fact that most isolates in those studies, were from admitted military personnel during Iraq and Afghanistan wars. This shows that, an increase in the use of broad-spectrum antibiotics along with war and natural disasters contribute to the isolation of more antimicrobial resistant bacteria. The current study also, describes the important role of class D carbapenem hydrolyzing β-lactamases, and in particular blaoxa-23-like gene, in the dissemination of imipenem resistant A. baumannii isolates in Tehran hospitals. 56% of A. baumannii isolates which possessed bla oxa-23-like genes, were resistant to both imipenem and meropenem, and 37 (86.04%) isolates of MDR A. baumannii possessed only bla oxa-23-like genes. Overall rate of resistance to imipenem was 83 (67.47%) and to meropenem 104 (84.55%).
Several reports from around the world indicate a large increase in the rates of carbapenem-resistant A. baumannii from 8% in 2003 to 52% and 74% in 2005 and finally to 96% in 2007 (17, 18). In Iran it was reported as 49.3% resistance to imipenem and 50% resistance to meropenem in 2008 (12), 52.5% resistance to imipenem and meropenem in 2009 (19), and 49.26% resistance to imipenem in 2011 (20). Distribution of blaoxa alleles among Acinetobacter isolates, in Tehran was as follow: blaoxa-23-like / bla oxa-51 like was detected in 25%, blaoxa-24-like / bla oxa-51-like in 17.9% and blaoxa-58-like / bla oxa-51–like was detected in 9% of the isolates in 2008 (12). Bla oxa-23-like in 25%, bla oxa-58 -like in 21.2% and bla oxa-24-like in 15% were detected in 2009 (12, 19).
In other studies, increasing level of blaoxa-23 was reported so that,94% and 84% of the isolates were positive for bla oxa-51 and bla oxa-23 like genes in 2011(20). 88.7% bla OXA-23-like, 1.6% bla OXA-40-like, and 3.2% had bla OXA-58-like resistance genes in 2012 in North West of Iran (21). The current study report in 2010-2011 also demonstrates an increased prevalence of bla oxa-23-like gene (81.3%), but different data about blaoxa-24-like (8.13%) and bla oxa-58 genes (0.81%). Explanation of this difference is that in one of the previous studies (12), all isolates of Acinetobacter (A. baumannii and NON-A. baumannii), sensitive and resistant strains were included. A part of this phenomenon may be due to the fact that different hospitals were evaluated.
It is expected that different hospitals present different molecular epidemiology of carbapenem-resistant. Studies in various parts of the world revealed considerable geographical differences in the types of class D carbapenem hydrolyzing β-lactamases (18). In Taiwan, in 2006, 45% of A. baumannii isolates were resistant or intermediate to imipenem and meropenem. However, they found only one bla OXA-23 and one bla OXA-24 harboring A. baumannii isolate (22). Similar observations have been reported in other Taiwanese studies (23, 24). In other countries, the widespread dissemination of carbapenem -resistant Acinetobacter spp. with bla oxa-23-like or bla oxa-24-like has been reported.
Mendes et al., during 2006 – 2007, from 41 medical centers located in 10 countries, reported the class D carbapenemase genes in 70% of the strains. Bla oxa-23-like was the most common gene, which accounted for 95.0% of the class D carbapenemase-encoding genes detected, followed by a lower occurrence of bla oxa-58 (11.9%) and bla 0xa-24/40 (5.6%) (18, 25). In Ohio, the United States in 2009, 13% of imipenem resistant isolates, contained the blaoxa-23.The other class D carbapenemase, including bla oxa-24 and bla oxa-58 like genes could not be identified (2). In Bulgaria, 72.72% of carbapenem-resistant isolates were positive for bla oxa-23-like and 27.27% were positive for bla oxa-58-like (17).
The current study found four strains that contained only the bla oxa-51-like. Three of these isolates were sensitive to imipenem and meropenem. Therefore it implies that the relationship between bla oxa-51-like and resistance of A. baumannii to carbapenem is dependent on other factors such as the presence of ISAba1-bla oxa-51-like that play an important role, as a ‘mobile promoter. It is suggested that the presence of ISAba1-blaoxa-51-like be examined. In addition, more studies are needed to determine the clonal relatedness to know if dissemination of the blaoxa genes results in strains that are derived from a common ancestor or the result of strains exchanging a transposable genetic element. It is also possible that the MDR strains which circulate in hospitals are distinct lineages or groups of lineages within A. baumannii, which suggests that the problem of resistance might be associated with a few numbers of A. baumannii lineages.
The current study showed low susceptibility rates to most of the available antimicrobial agents for treatment of infections caused by A. baumannii, except for polymyxin B and colistin, while other studies in Iran have demonstrated 12% resistance to colistin and 3% resistance to polymixin B in 2011 (20),and 8.8% resistance to polymixin B in 2009 (19), multi-drug strains resistant even to colistin suggests, we should be looking for novel Therapeutic strategies. It should be noted that, fight against MDR A. baumannii (and other MDR organisms) is far beyond the hospital and needs a common strategy of decision makers and health-care officials, the challenge being to make hospitals as a safe place for patients.