1. Background
Klebsiella pneumoniae (K. pneumoniae) is one of the most common Gram-negative bacteria showing resistance to multiple antibiotics Worldwide (1). Since the 1980s, extended-spectrum β-lactamase (ESBL)-producing Gram-negative bacteria have been isolated in many countries (2), and infections by ESBL-strains cause increased morbidity and mortality (3). These are a group of enzymes that are capable of hydrolyzing the third-generation cephalosporins, such as cefotaxime, ceftazidime, ceftriaxone, and the monobactam group of antibiotics, such as aztreonam, thus causing resistance to these antibiotics in ESBL-producing bacteria. ESBL is predominantly found in Klebsiellae spp. and Escherichia coli, and other members of the Enterobacteriaceae (4). The most prevalent ESBLs are included in three groups: TEM, SHV and CTX-M (5).
The majority of ESBLs identified in clinical isolates to date, have been SHV or TEM types, which have evolved from narrow spectrum β-lactamase such as TEM-1, TEM-2, and SHV-1 (6). Genes encoding these enzymes are located on transferable plasmids that often carry other resistance factors, including resistance to aminoglycosides (7). The first plasmid-mediated β-lactamase in Gram-negative bacteria, TEM-1, was described in the early 1960s. TEM-1 is able to hydrolyze penicillins and early cephalosporins (8). Another common plasmid-mediated β-lactamase that is found in K. pneumoniae and E. coli is SHV-1 (for sulphydryl variable) (9). Members of the SHV family of β-lactamases trace their descent to SHV-1, a plasmid-encoded β-lactamase that confers to K. pneumoniae high levels of resistance against ampicillin (10).
Additionally, a new series of ESBL enzymes, cefotaximases (CTX-M), resulting in higher MICs of cefotaxime and ceftriaxone than of ceftazidime, has been discovered in several members of the Enterobacteriaceae family and in various countries (11). The first CTX-M-type β-lactamases were identified as plasmid-encoded enzymes in clinical isolates of Entrobacteriaceae (12). CTX-M-type β-lactamases are increasingly becoming the predominant ESBLs globally in the recent years, as opposed to the conventional TEM and SHV-Type ESBLs (13). In contrast with TEM and SHV ESBLs, most of the CTX-M enzymes preferentially hydrolyze and confer resistance to cefotaxime and ceftriaxone rather than ceftazidime (14). CTX-M type ESBLs show only 40% identity to TEM or SHV ESBLs, but they are closely related to β-lactamases of the Kluyvera spp. (15).
2. Objectives
The aim of this study was to determine the prevalence of K. pneumoniae encoding genes for CTX-M, TEM-1 and SHV-1 ESBL enzymes isolated from clinical specimens in the University teaching hospitals, Ahvaz, Iran.
3. Material and Methods
3.1. Bacterial Isolates
In an 11-month study from August 2011 to July 2012, 500 Entrobacteriaceae isolates were collected from patients in Golestan and Razi teaching hospitals, Ahvaz, Iran. After immediate transferring of isolates to the laboratory of Infectious and Tropical Diseases Research center of the university, they were examined by traditional culture and biochemical identification tests (16). The confirmed clinical isolates of K. pneumoniae were stored in Tryptic soy broth (HiMedia, India) + 15% glycerol at the temperature of -70°C.
3.2. Antimicrobial Susceptibility Testing
Antibiotic susceptibility testing was performed by disc-diffusion method on Mueller Hinton agar medium (Hi Media, India) and the results were interpreted according to the current guidelines of Clinical Laboratory Standards Institute [CLSI] (17). The tested antibiotic discs were: Ampicillin (10 μg), Amikacin (30 μg), Amoxicillin (25 μg), Amoxicillin/clavulanic acid (20/10 μg), Piperacillin (100 μg), Cefuroxime (30μg), Piperacillin/tazobactame (100/10 μg), Cefotaxime (30 μg), Ceftazidime (30 μg), Cefepime (30 μg), Ceftriaxone (30 μg), Cefoxitin (30 μg), Ciprofloxacin (5 μg), Aztreonam (30 μg), Gentamicin (10 μg), Imipenem (10 μg) and Meropenem (10 μg). All the antibiotic discs were purchased from Mast Co., UK. E. coli ATCC25922 (β lactamase negative) and K. pneumoniae ATCC700603 (ESBL positive) strains were used as controls of the susceptibility testing.
3.3. Screening of ESBLs With Combination Disc Method
In brief, pairs of discs containing cefotaxime (30 μg) and ceftazidime (30 μg) with and without clavulanic acid (10 μg), were placed on opposite sides (at a distance of 20 - 30 mm) of one inoculated plate containing Muller Hinton agar. A positive test result was defined as a ≥ 5 mm increase in the zone diameter compared to a disk without clavulanic acid (18).
3.4. PCR for Detection of Involved Genes
DNA was extracted from the bacterial colony by simple boiling method. Using template DNA, the polymerase chain reaction technique (PCR) was used to detect SHV-1, TEM-1 and CTX-M-1 genes. Primers used to amplify the screened genes are listed in Table 1 (19 ).
| Genes | Primer Sequence |
|---|---|
| bla SHV-1 | F:5'ATGAGTATTCAACATTTCCG 3' |
| R:5' CCAATGCTTATTCAGTGAGG 3' | |
| blaTEM-1 | F:5' CTGGGAAACGGAACTGAATG 3' |
| R:5' GGGGTATCCCGCAGATAAAT 3' | |
| blaCTX-M-1 | F:5' GACGATGTCACTGGCTGAGC 3' |
| R: 5'AGCCGCCGACGCTAATACA 3' |
DNA amplification was performed in an Eppendorf thermal cycler (Roche Co., Germany) in a final volume of 25 µL containing 2.5 µL of 10X PCR buffer, 0.8 mg/µL MgCl2, 200 µM of dNTP mix, 10 pM of each primer, 0.5 U of Taq polymerase and 5 µL of the template DNA. PCR conditions for the SHV-1 gene comprised an initial denaturation step for 5 minutes at 95°C, followed by 32 cycles of 94°C for 1 minute, 57°C for 1 minute and 70°C for 1 minute, with a final extension step at 72°C for 10 minutes. For TEM-1, the amplification cycle consisted of 5 minutes at 95°C, followed by 30 cycles of 94°C for 30 seconds, 55°C for 1 minute and 72°C for 1 minute, with extension at 72°C for 10 minutes. For CTX-M-1, the amplification cycle consisted of 5 minutes at 94°C, followed by 30 cycles of 94°C for 30 seconds, 55ᵒC for 1 minute and 72°C for 1 minute, with extension at 72°C for 10 minutes.
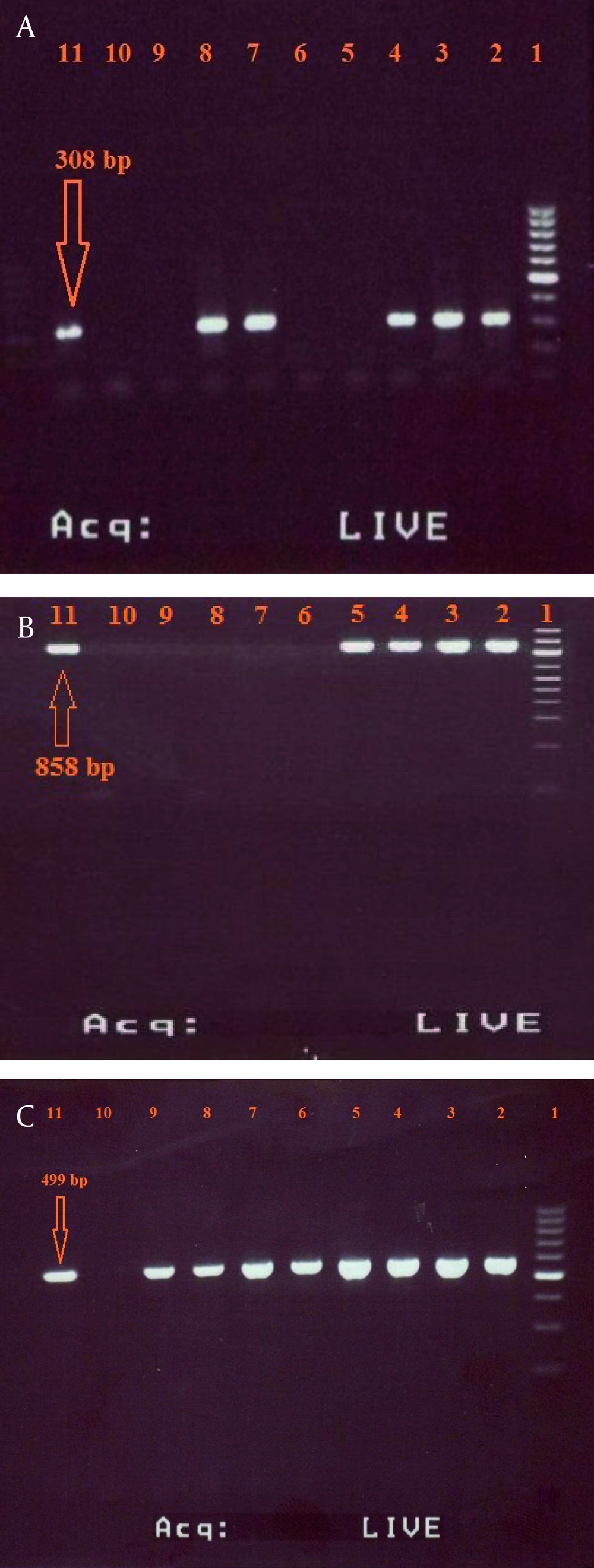
The primers used in the abovementioned reactions can amplify the DNA fragments correspond to 308 base pair (bp) of SHV-1 gene, 858 bp of TEM-1 gene and 499 bp of CTX-M-1 gene. K. pneumoniae strain ATCC700603 and E. coli ATCC25922 were used as positive and negative controls, respectively. The PCR products were loaded on a 2% (w/v) agarose gel with 0.5 mg/mL ethidium bromide and were analyzed by gel electrophoresis. SPSS software (SPSS Inc no. 13) was used for data analysis.
4. Results
In this study, among 500 clinical isolates of Entrobacteriaceae tested, 55 (11%) were identified as K. pneumoniae. These isolated were from 12 male (21.82%) and 43 female (78.18%) patients. The majority of the isolates were obtained from urine samples [39 (70.9%)] and the remaining [14 (29.1%)] from wound discharge.
Based on the result of combination disc method, 26 (47.27%) of K. pneumoniae isolates were ESBLs positive and 29 (52.73%) were ESBLs negative. According to the primary antimicrobial susceptibility testing, which is represented in Table 2, the majority of ESBL positive isolates showed high resistance to most of the tested antibiotics with highest rate of resistance to amoxicillin-clavulanic acid (100%), followed by amoxicillin (97.89%) and ampicillin (96.36%). Data analysis indicated that carbapenems (imipenem and meropenem) showed higher antibacterial activity against K. pneumoniae. The results of PCR method showed that the prevalence of SHV-1, TEM-1 and CTX-M-1 genes among ESBLs-positive isolates were 12 (46.15%) [Figure 1a], 9 (34.61%) [Figure 1b], and 7 (26.92%) [Figure 1c], respectively. furthermore, both genes of TEM-1 and SHV-1 were seen in four of the isolates (15.38%).
| Antibiotic | Susceptible, % | Intermediate, % | Resistant, % |
|---|---|---|---|
| Amoxicillin | 0 | 2.91 | 97.89 |
| Amoxicillin/clavulanic acid | 0 | 0 | 100 |
| Ampicillin | 1.82 | 1.81 | 96.36 |
| Amikacin | 9.1 | 25.45 | 65.4 |
| Aztreonam | 40 | 14.54 | 45.46 |
| Cefepime | 52.73 | 5.45 | 41.82 |
| Cefotaxime | 23.64 | 14.54 | 61.82 |
| Cefoxitin | 0 | 5.45 | 94.55 |
| Ceftazidime | 0 | 12.73 | 87.27 |
| Ceftriaxone | 36.36 | 18.18 | 45.46 |
| Cefuroxime | 0 | 10.91 | 89.09 |
| Ciprofloxacin | 29.09 | 38.18 | 32.73 |
| Gentamicin | 12.73 | 36.37 | 50.9 |
| Imipenem | 63.63 | 21.82 | 14.55 |
| Meropenem | 49.1 | 20 | 30.9 |
| Piperacillin | 1.82 | 23.63 | 74.55 |
| Piperacillin/tazobactame | 1.82 | 20 | 78.18 |
5. Discussion
K. pneumoniae strains producing various types of ESBL enzymes have spread worldwide. Among β-lactam molecules, carbapenems (imipenem, ertapenem, and meropenem) are the drugs of choice for treating infections by ESBL-producing Enterobacteriaceae (20). In this study, imipnem and meropenem were the most susceptible antibiotics against K. pneumoniae isolates (63.63% and 49.1% respectively). Though the first imipenem resistant strain isolated from a wound surgery was reported from Iran by Feizabadi et al. (21), however, imipenem was shown to be the most effective antibiotic against these isolates, as reported by another study from Iran (22).
Amoxicillin-clavulanic acid was the first β-lactam/ beta lactamase inhibitor combination approved for use in clinical practice, and is predominantly used as an oral preparation, effective against penicillinase- producing bacteria including E. coli, Klebsiellae spp, and P. mirabilis (23), although we noticed that all isolates were resistant to amoxicillin-clavulanic acid (100%) which is probably due to the frequent use of this antibiotic to treat infections caused by the members of Enterobacteriaceae (24). Such rate of resistance was also reported from Malaysian hospitals (25).
In this study, the rate of ESBL-producing K. pneumoniae was 47.27% based on the results from the combination disc method. This rate was in concordance with a report from Latin America (45.4%), though, it was higher than reports from Europe (22.6%), United States (7.6%) and Canada (4.9%) (26). In the Middle East, the prevalence of ESBL-producing K. pneumoniae in some parts has been reported as follows: Saudi Arabia 12.2% (27), Egypt 37.5% (28), Lebanon 20.0% (29), Turkey 50.0% (30), Jordan 80% (31), and Pakistan 36.0% (32). The rate of such strains is reported differently from various parts of Iran. Higher rates of isolated ESBL-producing K. pneumoniae are reported 77% by Mehrgan et al.(22), 72.1% by Feizabadi et al. (33), , 52.5% by Aminzadeh et al. (34), and the lower rates are reported 12% by Behroozi et al. (35), and, 19.6% by Irajian et al. (36). The reason for such prevalence differences may be attributed to the different broad-spectrum antibiotics consumption in the hospital setting, overuse of antibiotics in the community together with a lack of attention to laboratory screening of ESBL production by clinical isolates as other investigators concluded (22).
In the present study, the most prevalent ESBL genes were SHV-1, followed by TEM-1. Although TEM-1 was defined as the most frequent gene among ESBL producers in 1980s and early 1990, nowadays there are reports presenting SHV-1 as the most prevalent gene in many parts of the world (37). Similar to our findings, the prevalence of SHV-1 and TEM-1 among ESBL-producing K. pneumoniae strains in the study of Eftekhar et al. were 43.14% and 35.29% which were close to our findings, however their reported rate of CTX-M-1 gene was higher (31.37%) compared to this study (38). In a recent study from Iran, ESBL-producing K. pneumoniae was reported as 30.5% with the rate of 57% for SHV-1and TEM-1 (39).
Data analysis revealed that there was no significant correlation between the presence of genes SHV-1, TEM-1 and CTX-M-1 and sex of patients or type of clinical samples (P > 0.05). It is necessary to mention that we did not perform sequencing as a preferred complementary method due to some limitations, as the sequencing and comparing with standard strain shows us the probable mutations and the mechanism of resistance.
In conclusion, the rate of ESBL-producing K. pneumoniae was 47.27% in the present study showing that these are common in our hospital setting with resistant to many classes of antibiotics, resulting in limited treatment options. The greatest resistance was observed against Amoxicillin-clavulanic acid (100%) and the lowest rate of resistance was observed for imipenem (14.55%).
Management of infections resulted from these organisms is difficult and more complex, particularly in severe cases. Therefore, conducting molecular and epidemiological studies will help in identifying various types of ESBLs and establishing the control equipment for such strains in order to prevent and reduce their spread.