1. Background
Human T_cell lymphotropic virus type 1 (HTLV-1) is an oncogenic Retrovirus endemic in certain areas of the world including South Japan, West Africa, Caribbean island, Melanesians, North and South America and parts of Iran (1-3). The prevalence of HTLV-1 infection for the general population in certain areas of Iran such as Mashhad and Sabzevar is estimated to be 2.12 % and 1.6%, respectively and in Neyshabour this is 7.2% for the referred population (4-7). Approximately 5% of the estimated 20 million HTLV-1-infected individuals are at risk of eventually developing HTLV-1-associated diseases (8) such as adult T-cell lymphoma/leukemia (ATL or ATLL ), a fatal disease with Median Survival Time (MST) of less than 1 year (9) and chronic progressive disorder, HTLV-I Associated myelopathy/tropical spastic paraparesis (HAM/TSP) (10).
Other non-neurological or inflammatory conditions have also been reported (with varying degrees of evidence) to be in association with HTLV-1 including arthropathy, Sjogren’s syndrome, thyroiditis, pneumonopathy, children dermatitis and uveitis (11). The treatment of aggressive ATL with currently available therapeutic agents remains disappointing. HTLV-1-infected cells and ATLL cells from patients are highly resistant to multiple pro-apoptotic stimuli (12).
The determination and monitoring of virus in vitro and its infectivity is crucial for the analysis of drug resistance and various steps of the virus replication cycle (13). For this purpose, many techniques have been developed to detect retrovirus infection, including end-point dilution and plaque assays, antigen detecting by ELISA, various PCR-based assays and reporter systems (14, 15). However the non-cytolytic property of HTLV-1 is difficult to follow in contrast to that of HIV-1, for which various useful techniques are available (end-point dilution and plaque assays). On the other hand, these methods are expensive (ELISA) or there is a lack of correlation between viral nucleic acid detection and the presence of infectious particles (Realtime PCR) (16, 17).
Therefore due to the non-cytolytic characteristic of HTLV-1, development of a measurable biology system based on reporter gene in order to monitor HTLV-1 infectivity is necessary. Reporter systems are versatile and valuable tools in studies of gene expression, replication, fusion, and many other items that are widely used in basic science, molecular biology and medicine. For retroviruses, reporter system assays have been engineered for expressing genes such as β-galactosidase (β-gal) (18) and GFP (green fluorescence protein of the jellyfish) (13). Though this is a simple, economical, and direct method yet it requires fixation of cells, exogenous substrates (b-gal), and or is much less sensitive than the luciferase activity assay (in case of GFP) (19).
Due to the advantages of high sensitivity, high dynamic range and lack of internal luciferase activity in most eukaryotic cells, using luciferase has grown dramatically in recent years (18). HTLV-1 LTR luciferase reporter (pGL2 and pGL3-basic) vectors have been widely used to investigate the function of Tax proteins and their interactions with cell signaling pathways (20, 21). However the fourth generation of reporter vector, pGL4, containing both prokaryotic and eukaryotic selection marker genes, has been codon optimized for more efficient expression in mammalian cells. Furthermore, it has been engineered to reduce the number of consensus transcription factor binding sites, reducing background and the risk of anomalous transcription (22).
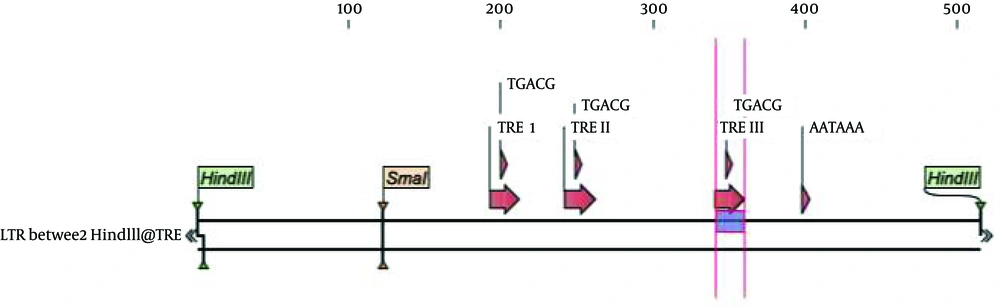
These methods usually rely on transactivation of the virus long terminal repeat (LTR) promoter. LTRs are located on both ends of the provirus, containing three regions U5, R and U3. In U3 there are three Tax respond elements (TREI, II and III) (23) which are similar in structure to the Cyclic AMP Response Element (CRE) and contain a core octanucleotide sequence. These regions are mostly responsible for Tax dependent transcription from provirus (24). The HTLV-I promoter is usually located in the 5´ LTR and is transactivated by the viral Tax protein. Tax, a viral oncoprotein, is a potent transcription transactivator that regulates HTLV-1 replication and facilitates the transition from quiescent infection to high levels of virus production in T-cells (25).
293T cell is a variant of HEK 293 cells (human embryonic kidney cells with sheared adenovirus 5 DNA) (26) that additionally contains the SV40 large T-antigen. It has been shown to be permissive to HTLV-1 infection (27).
2. Objectives
Although Iran is an endemic region for HTLV-1, very few papers are available for preparation of an indicator cell line for HTLV-1. Our aim in this project was to construct a reporter vector containing the luciferase gene coupled to the HTLV-1 promoter, in order to evaluate HTLV-1 infectivity in cell culture and aid future antiviral drug studies. The results have indicated success in making this vector.
3. Material and Methods
3.1. Cell Lines and Culture
293T cell line was obtained from the National Cell Bank of Iran (Pasteur institute of Iran, Tehran). This cell line was grown in high-glucose Dulbecco’s modified Eagle’s medium (Gibco, USA) supplemented with 10% heat inactivated fetal bovine serum (FBS), 100 units-100 µg/mL penicillin-streptomycin (PAA, Austria), and 2 mM L-glutamine, and kept at 37°C in 5% CO2. MT2 (HTLV-1-transformed human T-cell line) (28) and Jurkat (an HTLV unrelated human T-lymphoblast-like cell line) (29) were maintained in RPMI 1640 (PAA) with the same supplements.
3.2. Construction of Reporter Plasmids
3.2.1. Vectors
pUCLTR-lacZ has been described previously (30). It is a reporter plasmid, which carries b-gal reporter gene under the control of HTLV-I LTR. pGl4.17, a new generation of promoter-less reporter vector, was purchased from Promega Co, USA. pCDNA3.1Tax Hyg+ is an expression vector, containing a NF-kBless active Tax mutant, as previously described (31). pEGFP-N1 vector which carries enhanced green flourscence protein gene was purchased from BD Biosciences Clontech.(CA, USA). HTLV-1 LTR promoter containing three Tax respond elements (TRE I, II and III) (Figure 1) was cut out from pUCLTR-lacZ by Hind III digestion.
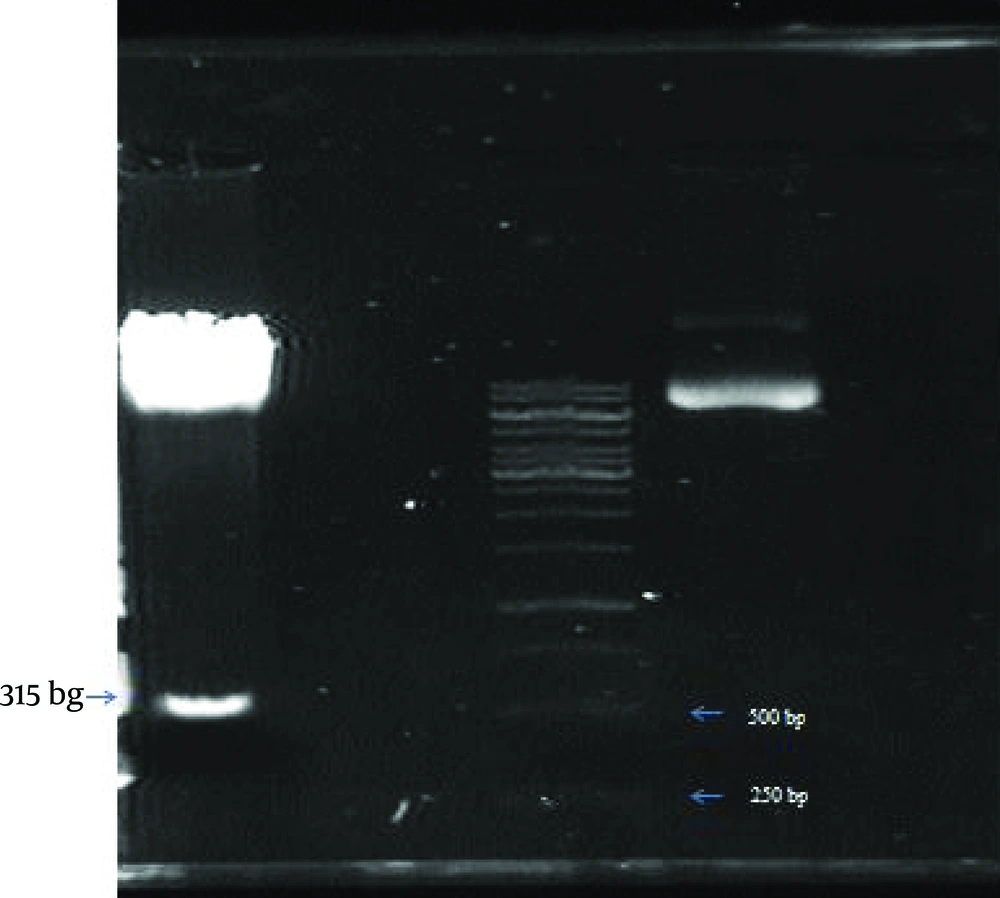
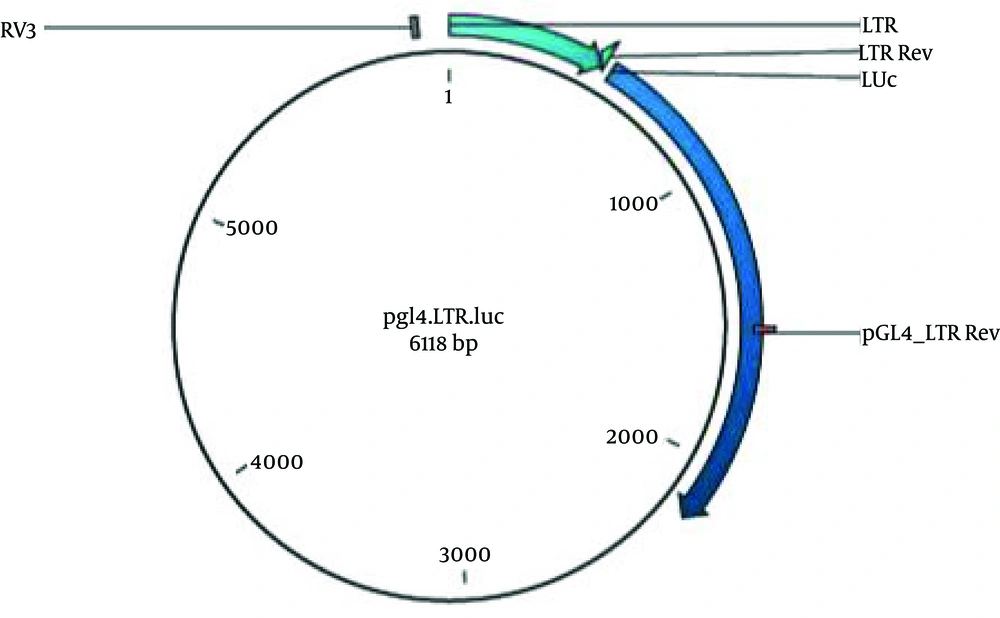
The 515 bp digested segment was purified from gel (Figure 2) (High Pure PCR Product Purification Kit, Roche, Germany) according to the manufacturer’s recommendation. LTR was sub cloned into the MCS (multiple cloning site) region of plasmid pGL4.17, upstream to the Luciferase reporter gene, using T4 ligase (Fermentas, Lithuania) to generate a pGL4LTR-Luc plasmid (Figure 3).
Competent Escherichia coli, strain DH5α was transformed by pGL4LTR-Luc, which was grown on LB agar in the presence of 50 µg/mL Amp. Colonies were screened by Colony PCR, using the Touchdown PCR protocol was performed with 20 cycles (95°C for 35 sec, 60°C for 40 sec with 0.5°C decrease per cycle for 10 cycles and 72°C for 75 sec), and universal RV3 (CTAGCAAAATAGGCTGTCCCC) and PGL4-LTR-Rev (TAGGTGGAAGCGTTTGGC) were used as forward and reverse primers. Once, selected colonies were grown in LB broth overnight, recombinant vector was purified by the QIAGEN Plasmid Mini Kit (Qiagen, Germany). Recombinant vector was confirmed by HindIII digestion and orientation of LTR was verified by double digestion (SmaI /XbI) and sequencing. This plasmid contains the G418-resistance gene that confers neomycin resistance to the transfected cells. Also pCDNA3.1Tax Hyg+, an expression vector, was used for induction of Luc expression.
3.3. Transfection and Luciferase Assay
The 293T cells were plated at a density of 8 × 104 cells per well in 24-well plates and allowed to grow overnight. 0.4 µg pGl4-LTR Luc plasmid was co-transfected with or without a Tax expression vector, pCDNA3.1Tax Hyg+, into the 293T cells using Polyfect (Qiagen, Germany), according to the manufacturer’s instructions. At 8 hr post-transfection, medium was replaced with fresh medium. After 48 hours, luciferase expression was performed using the One Glo system assay (Promega Inc., USA) on Synergy4 luminometer (Bioteck, USA). Quantilum (Promega, USA), a commercial luciferase, was used as a positive control. (Promega) Transfection conditions were normalized using pEGFP-N1 vector (Clontech, CA USA).
3.4. Co-culture of MT2 With Transfected Cell
In the same way 293T cell was transfected with pGl4-LTR Luc plasmid then co-cultured with MT2 (an HTLV-1 producer cell line) or Jurkat cells (as a negative control cell) and finally luciferase expression was assayed using the aforementioned protocol.
4. Results
4.1. Cloning Evaluation
Given that there is a chance for self-ligation of pGL4.17 during the ligation reaction which may enter into the bacteria and be capable to confer growth potential in the presence of ampicillin, screening was done on 80 colonies by Colony PCR. Among them, 32 colonies had LTR fragments that were inserted into the pGL4.17 vector. Besides, both ends of the LTR had been cut with the same enzyme (Hind III), making it likely for some plasmids to contain the inverted orientation. To check the accuracy of the LTR orientation, plasmid extractions of selected colonies were double digested by SmaI /XbI, followed by sequencing of 2 confirmed colonies. Further analysis using BLAST showed recombinant vectors carrying HTLV-1LTR segment containing TREs.
4.2. Luciferase Expression
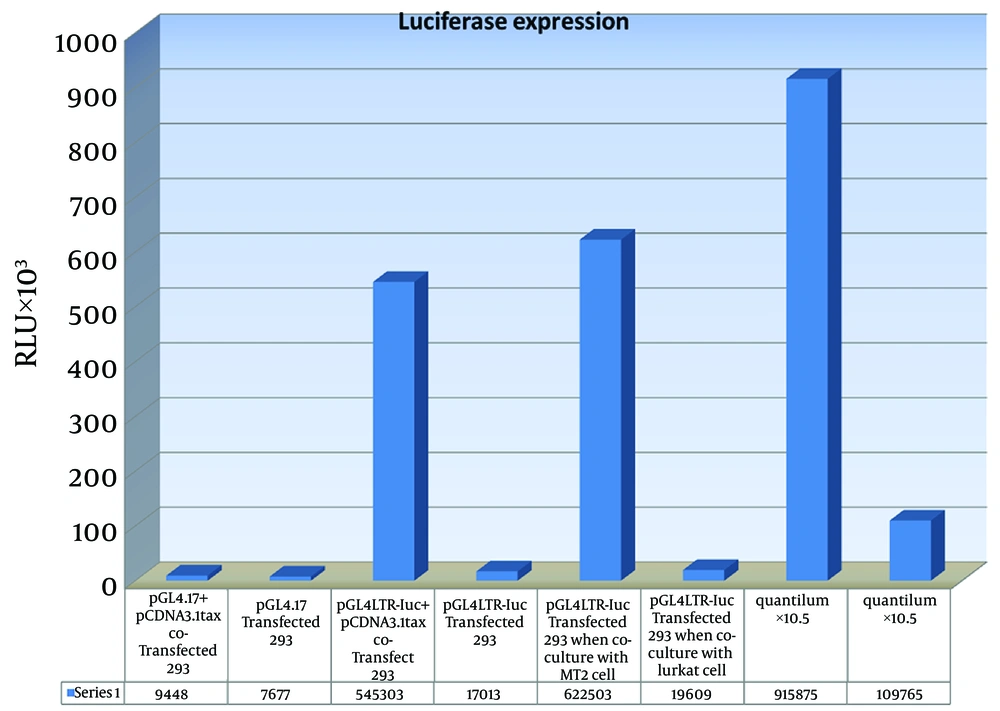
High levels of luciferase expression was seen when 293T cell was co-transfected with pGL4LTR-Luc, accompanied by pCDNA3.1Tax Hyg+, a Tax expression vector.In similar experiments, 293T cells were transfected with the pGL4LTR-Luc without pCDNA3.1 Tax Hyg+. Also promoter-less vector, pGL4.17, (as negative control) was introduced to 293T cell with or without pCDNA3.1Tax Hyg+.
The average luciferase expression was more than 50-fold higher in cells that were co-transfected with pCDNA3.1 + Tax and pGL4LTR-Luc than in cells that were transfected with pGL4.17 plasmid with or without pCDNA3.1Tax Hyg+. Average luciferase expression obtained from 293 T cells that were transfected with the pGL4LTR-Luc and co-cultured with MT2 cells, was more than 30 times compared to those co-cultured with Jurkat cells (Figure 4). Background of luciferase expression was negligible.
5. Discussion
There is no effective treatment for HTLV-1 linked diseases such as ATL and HAM/TSP and there is currently little research and development of new compounds with specific anti-human T-cell leukemia virus type 1 (HTLV-1) activity. It is clear that monitoring of virus in vitro and its infectivity is a crucial step for the analysis of drug susceptibility. Due to the non-cytolytic characteristic of this virus and its cell associated properties, there is no usual procedure for drug studies in vitro and it is limited to some HTLV infected cell lines (32). Hence, there is an urgent need to develop new strategies for drug discovery for these important diseases.
Reporter systems are instrumental and valuable tools that are widely used in basic science. There are several reporter genes which are used in bioassay systems such as Luc, GFP, CAT and β-gal. Among them luciferase has the most sensitive and high dynamic range. Moreover, cell lines have no auto luminescence. Many systems which have been reported mainly for retroviruses, have used similar strategies and also carry a reporter gene based on LTR Promoters (33). As Iran is an endemic region for HTLV-1 (7), here we made a luciferase reporter vector driven by the HTLV-1 long terminal repeat promoter. Thus the pGL4LTR-Luc responds to Tax expression during HTLV-1 infection by producing luciferase, a protein that can be readily and accurately quantitated in a luminometer.
In this study we showed that Induced expression of luciferase in 293 cells by Tax expression vector was more than 50-folds compared to the negative control while Azadmanesh and his colleagues (30) have declared that their reporter system induced a 10 times greater expression than the control. Their system was based on b-gal expression, so we believe that the difference is due to higher sensitivity of luciferase compared to ჩ-gal. In addition, although, qualitative assessment was the advantage of their system, yet fixation and staining procedures were necessary and were the disadvantages of their study compared to our study. In our study assessment was easily done only by cell lysis and required fewer cell enumeration.
However, the promoter of the virus can be affected by cell signaling pathways, considering the similarity of CRE with TRE (as previously was mentioned) (17). Background expression in this study was very low and thus negligible in contrast to the previous report. Nonetheless, more than 30 times difference was seen between the test and background. A similar result was obtained when 293T cell was transfected with pGL4LTR-Luc and co-cultured with Jurkat cells (an HTLV unrelated human T-lymphoblast-like cell line). For HTLV-1, the human kidney cell line, 293T, has been shown to be permissive to infection (27). We used the 293T cell line, to evaluate our construct in cell culture so that we could eventually establish an indicator cell line for subsequent studies.
The results obtained in this study are in line with Yoshida et al. studies. They have designed an indicator cell lines for HIV-1, and by the same strategy, they have established an indicator lymphocytic cell line for HTLV-1 replication carrying luciferase gene as a reporter. They have mentioned that this indicator cell line was insensitive to infection with cell-free HTLV-1 but observed induction of luciferase expression by Tax more than 40 times. They used third generation reporter vector, pGL3, that has no selection marker and needs to be co-transfected with a vector that provides an antibiotic gene to establish an indicator cell line; however, our construct consisted of forth generation Promega reporter vectors with resistance gene of neo.
In HTLV-1 infection, viremia is essentially a cytoviremia, since the virus is cell associated. Other Deltaretroviruses titers are known to be low both in culture and in their hosts. Jewell (27) compared the effect of Tax expression plasmid versus HTLV-1 producer cell line, MT2, to induce an indicator cell expressing GFP coupled to LTR. He found that the percentage of GFP-expressing cells were considerably higher (two- to four-fold) when transfected with the HTLV-1 Tax expression plasmid compared to levels observed when the same cells were co-cultured with MT-2 cells. He explained that the difference was most likely due to the low infectivity of virus from the MT-2 cells. Here, our construct was tested with HTLV-1 producer cell line, MT2, and Tax expression plasmid. Levels of luciferase expression that were induced in both experiments were higher than in Jewell’s experiment (for example more than 50 versus 14 folds). In spite of using the same cell line (293T), our construct had greater sensitivity because we used Luciferase as a reporter instead of GFP. Given the high functionality of the produced recombinant vector, it seems to be a good applicable tool for making an indicator cell line for future basic and drug discovery studies.