1. Background
Infectious diseases are common all over the world. A recent World Health Organization (WHO) report indicated that the infectious diseases are now the biggest death cause for children and young adults in the world. It has recently been reported that infectious diseases are responsible for more than 17 million deaths worldwide each year (1), among which two million are caused by water borne pathogens, (2) mostly associated with bacterial infections. Hence, the control of water borne infectious diseases is still a concerning issue worldwide. The ability to control such bacterial infections is largely dependent on using accurate and simple detection methods for ethological bacterial species in the clinical microbiology laboratories. Most developing and developed countries fail in detecting novel pathogenic water borne bacteria, while it is an epidemiologic concern to detect them in the very early stage. Water sources are over contaminated by sewage and pathogenic agents as well as industrial, agricultural and trade wastes. Water borne diseases have major impacts on the public health (2).
Numerous hospital outbreaks, mostly Acinetobacter sp. and Bacillus sp (1, 3) related infections, have been reported in the literature. In a small number of outbreaks the source of isolated strain from clinical specimens was taps; (3, 4) but identification of those bacteria is very difficult. By isolating microorganisms from pure cultures and detecting their morphological and physiological properties, microbiologists have been able to identify, characterize, type and quantify a wide range of microbial species. Remarkable advancement in microbial diagnosis, particularly molecular identification of pathogenic bacteria is very important. For successful identification of organisms in the genus or even more frequently in the species level, modern clinical microbiology diagnosis combinational methods of colonial morphology, physiology and biochemical or serological markers can be functional (1). Correct epidemiological reportage (5) of causal agents as well as accurate identification of microorganisms are crucial in a given disease state as our criteria, to develop a simple genomic DNA isolation method from cultivable bacteria protocols using a single chemical substance.
The techniques based on isolation and further biochemical or immunochemical characterization have been used for microbial categorization. From the present point of view, all these methods are complicated and ambiguous. Possessing simple materials without any conventional and large chemical procedures is very important for identification. The basic steps of genomic DNA isolation are: breakage of the cellular structure to create a lysate, separation of the soluble DNA from the cell debris and other insoluble materials and purification of the target DNA from soluble proteins and other nucleic acids by ethanol precipitation (4). These methods are time consuming and use a variety of dangerous reagents; using large chemicals can affect the efficiency of the method.
Ethanol, Hydrogen peroxide and phenol (6-8) are the common substances that specifically act on the cell membrane, inactivate the intra-cytoplasmic enzymes and denature the cell membrane proteins (9), followed by formation of unstable complexes which lead to the lysis of all gram positive and gram negative bacterial cell membranes (10). Genomic DNA is very useful not only for PCR manipulations but also for gene level identifications. Bacterial 16S rRNA genes contain nine hyper variable regions (V1–V9) that demonstrate considerable sequence diversities among different bacteria (11). Species-specific sequences within a given hyper variable region are useful targets for diagnostic assays and other scientific investigations, since no single region can differentiate all existing bacteria.
2. Objectives
Genomic DNA is important for identification of novel genes and diversity assessment of microbes in different habituates. The present work reports successful DNA amplification as well as identification of clinical important pathogens (1) such as Gram positive Bacillus sp. and Gram negative Acinetobacter sp., using 16s rRNA amplification and novel genomic DNA isolation methods (8). Sample preparation platform has been developed for rapid and simple detection of infectious organisms from the care diagnosis point and molecular manipulation has been performed using three novel genomic DNA isolation protocols. Prompt availability of genomic DNA (7) from microorganisms is necessary for gene cloning and selecting recombinant constructs, for taxonomic and diagnostic purposes.
3. Materials and Methods
3.1. DNA Isolation From Bacterial Culture
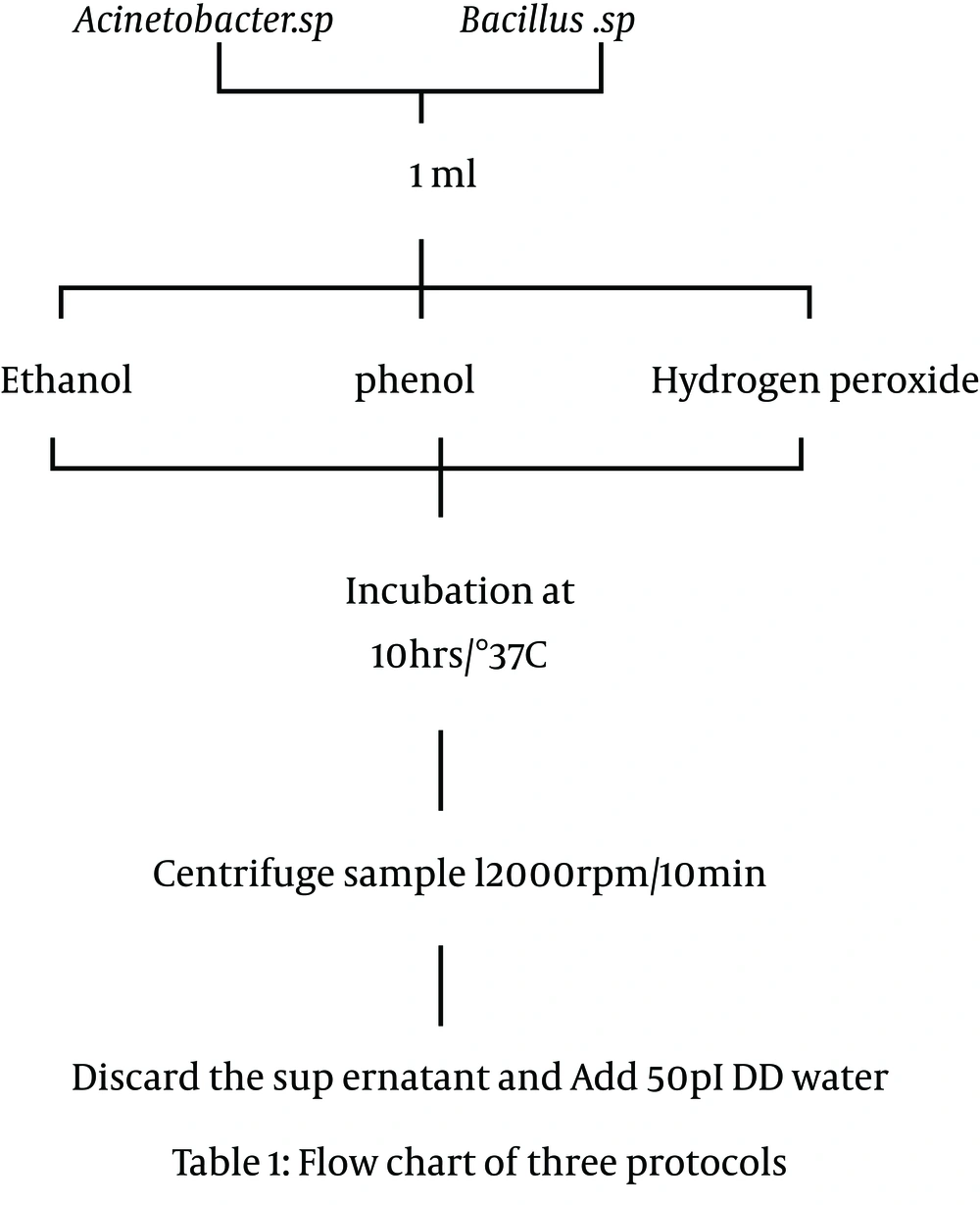
Two medically relevant species were used to assess the efficiency of three DNA extraction protocols. The panels of pathogenic bacteria, considered as experimental models to describe the novel protocols, were Bacillus sp. and Acinetobacter sp. Which were isolated from a mixture of water samples collected from Makthal and Nagarkarnool regions located in Mahabubnagar District, Andhra Pradesh, India. Unpurified drinking water samples were incubated overnight. The day after, selected Bacillus sp. and Acinetobacter sp. cultures were inoculated in two separate 1-mLeppendorfs of nutrient broth containing medium by the Actinomycetes and Bacillus cultures.
Smaller cultures were enough to achieve a minimum quantity of reproducible yield and the cell density was determined. The tubes were incubated overnight until the OD value was 0.5 at 590 nm for the cultures. Cell suspension was taken, spun at 6000 rpm for 4minutes and pelleted down. From each of 90% ethanol, 30% hydrogen peroxide and concentrated phenol,1 mL was added to each tube and incubated for 10 hours. The mixture was centrifuged at 12000 rpm for 10 minutes. Later, the supernatant was discarded so the pellet contained high molecular weight DNA. The efficiency of cell lysis was also measured by acridine orange method. The DNA was purified using the phenol-chloroform organic exaction method, together with enzymatic degradation of RNA contaminates using RNase (Figure 1).
3.2. PCR Analysis
The small subunit rRNA genes of cultured Acinetobacter sp. and Bacillus sp. DNAs were amplified using 16S rRNA universal primers. The 50 µL PCR amplification reaction mixture contained 4 µL of bacterial DNA (nearly 200 ng), 1 µL of Taq-DNA polymerase, 5 µL of Taq buffer, 5 µL of 2 mM dNTP mixture, 5 µL of forward primer (10 pM/µL) and 5 µL of reverse primer (10 pM/µL). Amplification was carried out in a Bio-Rad thermal cycler, run for 30 cycles. In each cycle, denaturation was done at 94ºC for 20 seconds, annealing at 48ºC for 20seconds and extension was done at 72ºC for 40 seconds, and a final extension was carried out for 5 minutes at 72ºC at the end of all 30 cycles. The amplified DNA fragment of approximately 1542 base pair (bp) was separated on a 1% agarose gel and purified using Quiagen spin columns.
The desired DNA band was cut from the agarose gel, weighed and then transferred to a sterile microfuge tube. Afterwards, QE buffer was added, thrice the volume of weighed excised band. The tube was placed on a thermo-mixer at 65ºC for 10 minutes and the contents were then transferred to a Quiagen column and spun at 8000 xg for 2 minutes. Then it was washed with 750 µL of PE buffer and eluted with a small quantity (30-40 µL) of sterile water. The purified PCR product was then used for sequencing.
3.3. 16S rRNA Gene Sequencing
The purified 1542 bp PCR product was sequenced using universal primers. The resultant, almost the complete 16S rRNA gene sequence of the isolate, was subjected to BLAST sequence similarity search to identify the nearest taxa. The entire related 16S rRNA gene sequence was downloaded from the database (http://www.ncbi.nlm.nih-gov) and aligned using the clustal–program.
4. Results
As a proof of the novel protocol of diagnosis and isolation for genomic DNA, we used Bacillus sp. and Acinetobacter sp. with questionable pathogenicity (3). These bacterial cultures are resistant to commercially available antibiotics due to possession of diverse resistance mechanisms (5). Pathogenic bacteria species exist in different locations of Mahabubnagar district, Andhra Pradesh, India. Acinetobacter sp. is an increasingly important healthcare-associated antibiotic-resistant pathogen, causing nosocomial infections and hospital water contamination (3). The diagnostic concerns of the bacteria with epidemiological and etiological pathogenic it yare noticeable in order to determine their molecular characterization (3).
Culture incubation time affects both the yield and quality of the isolated genomic DNA. DNA yield will be relatively low if the density of the bacterial cultures insufficient and vice versa (10). Successful isolation of high-quality genomic DNA begins with the culture preparation. A number of factors can influence the growth of gram positive and gram negative bacteria (11). Rigid bacterial cell walls are unusual disturbances in the disruption of many cells. In a bacterium, necessary living and reproduction procedures causes the cell to die. Particular protein folding is required for the protein function inside the cell. For the cell wall disruption, disinfectants are the common substances applied to non-living objects to destroy their living microorganisms. To maintain the chemiosmotic balance of a bacteria, the cell membrane normally acts as a diffusion barrier between the cytoplasm and the extracellular medium (6, 9). Ethanol is the only industrial material used as bactericide and fungicide but not as sporicide. Their mechanism of action appears to involve protein denaturation and membrane lipids dissolution.
Ethanol kills a bacteria (7) first by making the lipids -which are part of the outer protective cell membrane of each bacterium-more soluble in water so that the cell membrane begins to lose its structural integrity. Alcohol disrupts the folding of proteins which is called denaturation, so the protein loses its biological activity and can no longer function. One milliliter of 90% ethanol is enough to kill 80% of the bacteria and other microbes. At concentrations above 1%, phenol (6) has significant antibacterial effect. Derivatives of phenol, called phenolics, injure the lipid-containing plasma membranes of bacteria and cause the cell to leak its cellular contents as well as the genomic DNA. Peroxides play an important part in the bacterial cell degradation (6). Oxidized molecules are more sensitive to proteolysis than other molecules, and it has been suggested that a system of cell sanitization may be more effective together with an oxidizing agent such as H2O2 and result in an easier DNA extraction.
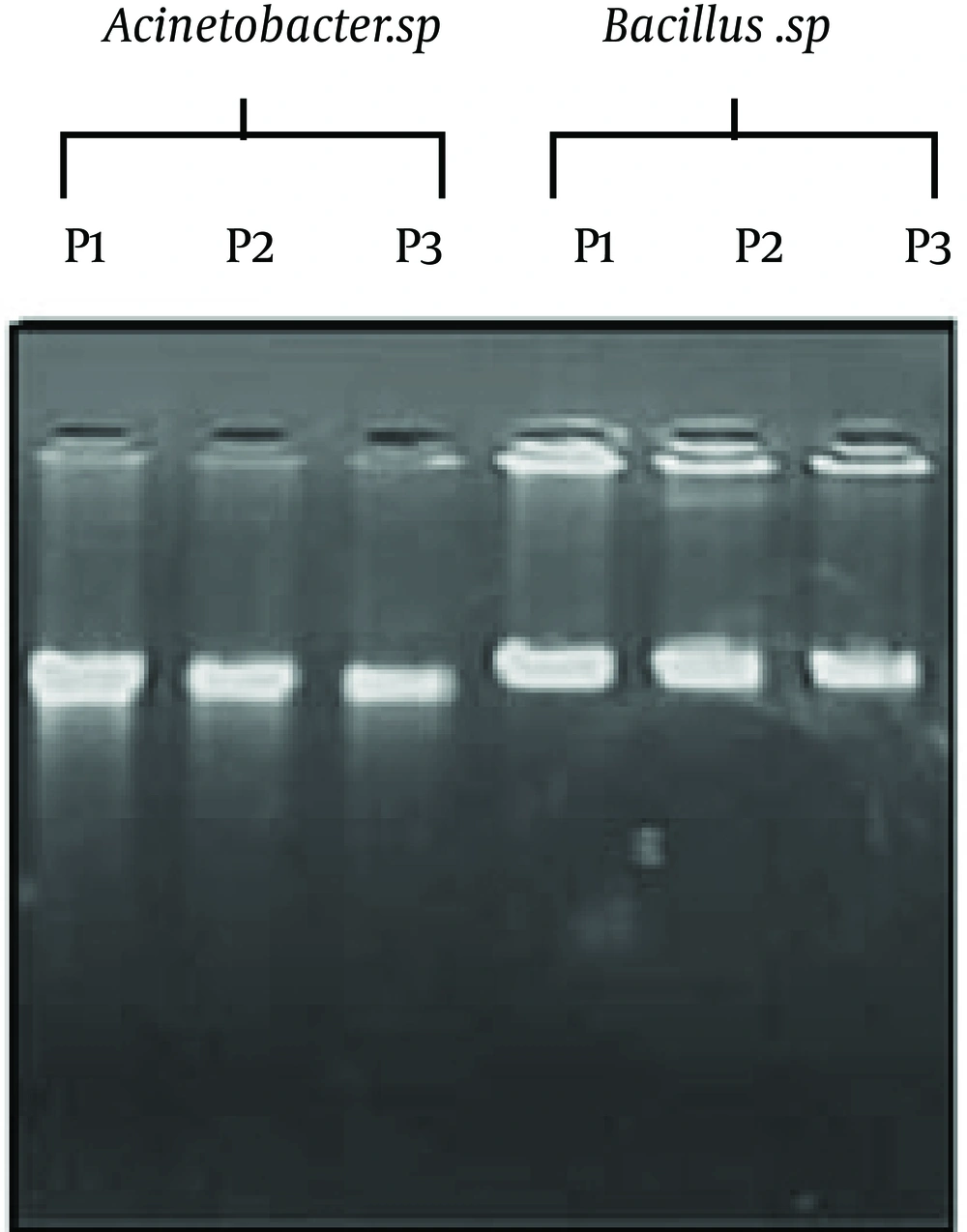
With reference of Figure 1, after 10 hours of incubation with 1mL of 90% ethanol, phenol and 30% hydrogen peroxide (9) for all the samples, they were continuously centrifuged at 10000rmp for 10minutes. The comparative effectiveness of the three present protocols is mainly based on the disruption of bacterial cell wall, genomic DNA yield and the manipulation capacity. Ethanol and phenol are the best agents, but for hydrogen peroxide, because of the formation of free radicals, genomic DNA may slightly denature. Furthermore, this did not show any effect on further DNA manipulations such as pathogenic bacteria detection and PCR (12). Figure 2 represents the agarose gel pulsed electrophoresis of genomic DNA of Acinetobacter sp.(set 1: P1, P2, P3) and Bacillus sp. (set 2: P1, P2, P3) using the protocols of ethanol, phenol and hydrogen peroxide. All the protocols resulted in good quality DNA, but the intensity of bands were slightly variant (Figure 3). Usually, ethanol and phenol are good materials for genomic DNA extraction, but in the hydrogen peroxide protocol, the band intensity was slightly lower than others.
5. Discussion
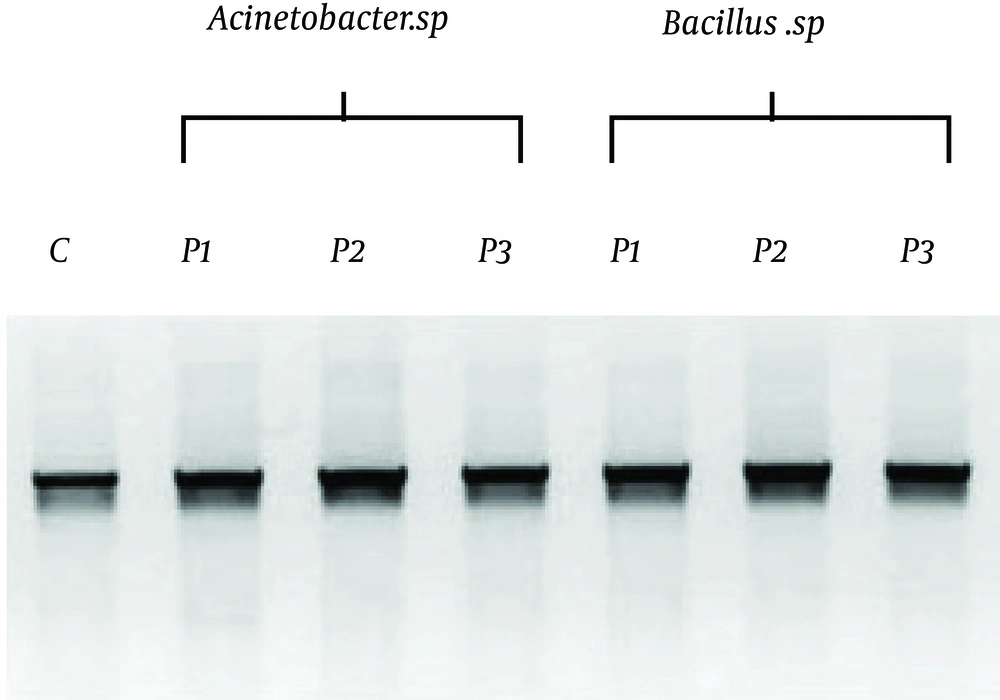
The focus of genomic DNA extraction methods has been on rapid function of molecular techniques, avoiding extensive purification steps (13,14 ). The described methods including PCR can successfully act from both ends to detection the novel pathogenic bacteria and their phylo genetic taxa. Acinetobacter sp. and Bacillus sp. were amplified using a single set of primers, forward and reverse, with a reaction mixture combination, and gel electrophoresis was run afterwards. Figure 3 represented three different novel amplification protocols for identification of two waterborne pathogenic bacteria, Acinetobacter sp. (first three lanes) and Bacillus sp. (second three lanes). After that, the genus level was identified by 16s rRNA sequencing and Acinetobacter sp. (GU 566361) and Bacillus sp. (GU 566359) were identified as the nearest taxa using the BLAST analysis.
Novel protocols have always had an authentic role in medical molecular biology and diagnoses of novel pathogenic bacteria. The present study on Acinetobacter sp. and Bacillus sp.is a proof that we can achieve high yields of genomic DNA and conduct a diagnostic study of novel pathogenic bacteria as well as their identification and amplification at the genus level.