1. Background
Helicobacter pylori (H. pylori) is a microaerophilic spiral-shaped Gram-negative bacterium, which is found in the gastric mucous layer or adherent to the epithelial lining of the stomach (1). The prevalence of H. pylori infection is reported as approximately 50%, worldwide, which is as high as 80%-90% in developing countries (2). Although most infected persons remain asymptomatic, but 15-20% of H. pylori- positive individuals will develop the associated diseases. The occurrence of diverse diseases as peptic ulcer, gastric cancer and MALT lymphoma with H. pylori depends on specific features of the organism, host genetic and environmental factors (3).
H. pylori express the virulence factors that augment the risk of clinical disease outcomes. The virulence factor that has attracted most of the interests is the cytotoxin-associated gene A (cagA), which encodes the CagA protein and the cellular effects of CagA may clarify why patients infected with cagA strains, usually have higher inflammatory responses and level of proinflammatory cytokines. Thus, these gastric mucosal responses induce the peptic ulcer diseases, precancerous lesions, and cancer in adults (4). Approximately 60% of H. pylori strains isolated in Western countries carry cag pathogenesity island (cag PAI), whereas almost all of the isolates from East Asia are cag PAI-positive (5).
The Maastricht Ⅲ consensus advised that proton pump inhibitors (PPI), plus clarithromycin, amoxicillin or metronidazole therapy for 7 to 14 days as the first choice for H. pylori infection management (6). Recently, several large clinical trials and meta-analyses have shown that the eradication of the standard triple therapy has generally declined to unacceptable levels (i.e., 80% or less) (7). Resistance to antibiotics is the chief factor for treatment failure (2). In other way, results of some studies have suggested that the eradication rate in patients with gastritis is lower than that the patients with peptic ulcer diseases (8). Since the worldwide increase of the drug resistance rates represents a problem of relevance, some researches have been conducted on antibiotic resistance relationship with bacterial genetic factors (9, 10). If harboring cagA affects the response rate of therapy, then it may be possible to anticipate H. pylori eradication rates.
2. Objectives
The aim of this research was to analyze the relationship between the antibiotic resistance and cagA presence in H. pylori isolates
3. Patients and Methods
3.1. Patients
One hundred and twenty three H. pylori isolates were recovered from gastric biopsy samples of patients with gastritis, peptic ulcer and gastroesophageal reflux diseases underwent endoscopy in Azerbaijan, Iran. At least 1 week prior to endoscopy the patients had stopped receiving antimicrobial agent.
3.2. Bacterial Isolates
For bacterial culture, the gastric biopsy samples were homogenized and cultured onto the Brucella agar (Pronadisa Co, Spain) containing 5% sheep blood and antibiotics supplement (vancomycin 6 µg/mL, amphotricin B 2.5 µg/mL and trimethoprim 20 µg/mL). The cultured plates were incubated under microaerophilic condition (Mart, Anoxomat, Lichtenvoorde, Netherlands O2 5%, CO2 10%) at 37°C with high humidity for 5 - 7 days. H. pylori was detected based on colony morphology, by Gram staining and positive oxidase, catalase and urease tests.
3.3. Antibiotic Susceptibility Tests
Antimicrobial susceptibility of H. pylori isolates were determined by modified disk agar diffusion test. Suspensions of 3-day old cultures were prepared in sterile normal saline to match with No. 4 McFarland standard (11). The disks of antibiotics (MAST Co., England) including clarithromycin (15 µg), amoxicillin (25 µg), and metronidazole (5 µg) were placed on the inoculated plates and incubated for 72 hours. The inhibition zones were interpreted as previously reported (12).
3.4. DNA Extraction
Genomic DNA was extracted with the CTAB reagent method (13). The extracted DNA eluted in 50 μL of 1 × TE buffer (10 mM Tris-HCl, 1 mM EDTA (pH 8.0)) and stored at -20°C until use.
3.5. Assessment of cagA and glmM Genes by Multiplex PCR
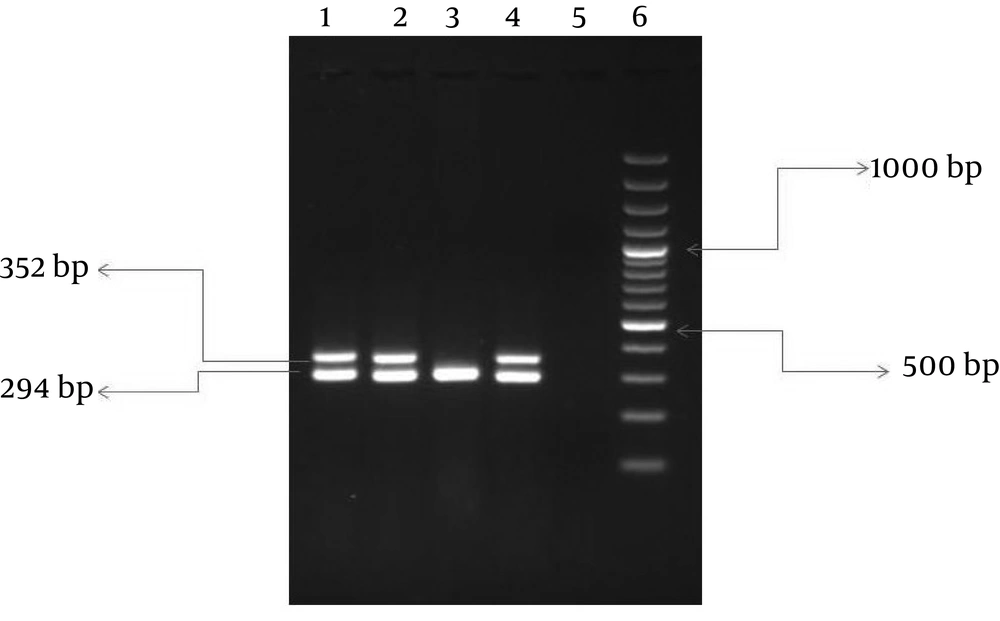
Primer sequences (Table 1) and conditions of PCR amplifications of the glmM gene for the detection and confirmation of H. pylori and the cagA virulent gene were designed based on previous studies ( 14 , 15 ). Briefly, each PCR was performed in a total volume of 50 µL containing 100 ng H. pylori genomic DNA, 0.2 µL of dNTP (10mM), 0.2 µL PCR buffer (1X), 0.1µL mgcl2 (1.5 mM), 0.2 µL of each primer and 1.5 µL of Taq polymerase (500U). The PCR was carried out in an automated thermal cycler (Eppendorf, Germany). After amplification, 10 µL of PCR product was electrophoresed onto 1.5% agarose gel stained with ethidium bromide and later visualized under UV illuminator.
| Gene | Primer | Nucleotide Sequence | Size (bp) |
|---|---|---|---|
| ureC (glmM) | HP-F | GGATAAGCTTTTAGGGGTGTTAGGGG | 294 |
| HP-R | GCTTACTTTCTAACACTAACGCGC | ||
| cagA | cagA-Fm | AGGGATAACAGGCAAGCTTTTGA | 352 |
| cagA-Rm | CTGCAAAAGATTGTTTGGCAGA |
3.6. Statistical Analysis
The correlations between the H. pylori-cagA genotype and antibiotic susceptibilities were analyzed using χ 2 test or Fishers exact test. A P value less than 0.05 was considered as statistically significant.
4. Results
The mean age of the patients was 35 ± 18 years. Out of 123 H. pylori isolates, 99 isolates were collected from adults, and 24 isolates from children. Overall, the rate of resistance to clarithromycin, metronidazole and amoxicillin were 17.07, 78.86 and 27.64%, respectively. Resistance to both metronidazole and clarithromycin were detected in 13% (n = 16). Of interest, 10 isolates (8.13%) were resistant to all antibiotics (clarithromycin, metronidazole and amoxicillin). The cagA gene was detected in 84 strains (68.3%) (Figure 1). The distribution of the cagA gene was the same in both genders and age groups (73% children and 68% adults, 68.6% male and 68.8% female).
The presence of cagA observed more likely in patients with peptic ulcer diseases (80%) than in non-ulcer dyspepsia ones (65%), and the difference was statistically significant (P value ≤ 0.05). The frequency of resistance to clarithromycin, metronidazole and amoxicillin in H. pylori - cagA positive were, 16 (13.01%), 67 (54.47%) and 24 (19.51%), respectively. While the frequency of resistance to these antibiotics in H. pylori - cagA negative were 5 (4.06%), 30 (24.39%) and 10 (8.13%), respectively. The distribution of cagA in different groups of gastroduodenal diseases and antibiotic resistance are demonstrated in Table 2.
a Abbreviations: Am, amoxicillin; Cl, clarithromycin; Met, metronidazole
5. Discussion
The three main antibiotics that are used as first-line eradication regimens against H. pylori are clarithromycin, metronidazole and amoxicillin (16, 17). However, H. pylori resistance to antimicrobials agents is the main cause of treatment failure (18). The level of resistance varies between different geographical regions and unfortunately, which is increasing worldwide. This study showed that the rate of resistance to metronidazole is high (78.86%). In developed countries, metronidazole resistance ranges from 10 to 50% while in developing counties, higher rate of resistance are reported (19). This is may be related to the greater consumption of this drug (2). In the case of clarithromycin, the second most commonly used antibiotic in H. pylori management, we found resistance rate as 17.07%. However, in Asian countries, a higher clarithromycin resistance rate has been reported in Japan (40.7%) while, the lowest value has been observed in Malaysia (2.1 %) (18). In North America, clarithromycin resistance ranges from 2.5 - 12.2 % (20).
In this research, the amoxicillin resistance was found in 27.64% of isolates. Until now, the resistance to amoxicillin in H. pylori was reported to be very rare; but have now been reported in USA (21), Italy (22), Brazil (23) and Iran (2, 24). Multiple drug resistance is a major problem worldwide. In the present study, we remarkably observed double resistance to clarithromycin and metronidazole and triple resistance to clarithromycin, metronidazole and amoxicillin. Pretreatment H. pylori antibiotic resistance has been reported to compromise the efficacy of management. For example, therapy regimens containing metronidazole and clarithromycin fail in as many as 38% and 55% of cases, respectively, when used to treat infection with an organism non-susceptible to one of these antimicrobial agents (17).
There are new facts about the existence of various strains of H. pylori with different levels of virulence (1, 3, 8, 25-27). The cagA gene is located at one end of a 40-kilobase DNA section called cag pathogenicity island (cag PAI) (28, 29). The clinical relevance of the putative virulence- associated genes of H. pylori and geographical region is still a subject of controversy (28). The prevalence of cagA-positive H. pylori strains varies from one geographic region to another (29). In this research such as the study from Isfahan- Iran (30), the prevalence of cagA-positive H. pylori strains was 68.3% which is more similar to the findings reported from Western countries (18, 27) and Brazil (31) than those reported from East or Southeast Asia (14, 28, 32). The presence of the cagA gene is thought to be associated with a further severe clinical outcome of the disease (3, 27, 33-35). The current study demonstrated the high prevalence of H. pylori infection containing cag A in patients from Azerbaijan with peptic ulcer disease than non-ulcer dyspepsia (Pv 0.02). Reports from Turkey (3) and Iraq (36) have shown similar results to our findings. Due to the allelic variation in cagA the H. pylori subtypes can circulate in different regions, the differences in cagA subtype might be a marker for different virulence among cagA positive H. pylori strains (32).
Furthermore, the severity of gastric inflammation and the pathology also affect the outcome of therapy (8, 9). There is now increasing evidences that cagA-positive strains induce an enhanced inflammatory response, mucosal damage and significantly delays healing of ulcers (1, 8). In this study, the rate of resistance to antibiotics in H. pylori- cagA positive were higher than cagA negative ones, however no statistically significant difference was found (P value > 0.05). Similarly, some studies also failed to find any general association between virulence markers and antibiotic resistance (18, 22, 37). This result is inconsistent with other studies that suggesting the statistically relation between drug resistance and cagA-positive strains (27, 31, 38, 39). There is little information about the relation between the variability of the strain and any outcome on the drug resistance rates of H. pylori. At the molecular level, there is no hypothetical basis for predicting an association between drug resistance and genotype; as such loci show to be neither physically nor functionally linked by genome organization (40).
Our research had some limitations. The studied population was not large enough, and our results need to be confirmed by studies on great number of patients. This study only tested the hypothesis that the cagA gene is related to antibiotic resistance. We think additional genetic or environmental factors might influence the interaction of H. pylori and antibiotics. In conclusion, the prevalence of H. pylori resistance is high in this area. Our results indicated that the relationship between drug resistance and the availability of cagA gene was not statistically significant. So, different treatment regimens are not recommended for patients with H. pylori- cagA positive infection.