1. Background
Low pathogenic H9N2 avian influenza viruses have caused outbreaks in poultry, resulted in serious economic losses in Asia. The Euroasian viruses grouped in three sub-lineages on the basis of antigenic and genetic properties (1). The direct transmission of H9N2 viruses from avian to human (2) and development of multiple reassortant H9N2 subtypes (3-5) introduce the virus as a potential pandemic candidate by the World Health Organization. From 1990s, many studies have been made to introduce suitable alternatives of eggs for vaccine production and research purposes. Recently, new cell lines are considered for supporting the replication of a wide variety of high virus strains titers (6, 7).
Alveolar epithelial cells and macrophages are the main targets for influenza virus (8). Some related factors to either virus or host involve in the efficient viral propagation in the cells. Hemagglutinin (HA), neuraminidase (NA), and nonstructural (NS) genes play important roles in host specificity and virulence of influenza virus by mediating viral entry into the target cells and membrane fusion (9), removing the sialic acid (SA) receptors and facilitating the release of progeny virus particles (10), and suppressing host innate immune responses (11). The identification of virus-host interaction is essential to investigate potential cell culture systems for explanation of virus replication and adaptation. The interaction depends on the ability of the virus to enter host cells followed by specific binding to SA receptors and activation of HA protease processing (12).
Generally avian influenza viruses bind to terminal α2,3 SA residues whereas human viruses bind to α2,6 SA. The changes in receptor specificity in H9N2 viruses lead to a higher affinity binding of α2-6Gal and SA found in the human upper respiratory tract (10). Lee et al. (13) have been suggested that H9N2 viruses belonging to G1 sub-lineage, may be better adapted to the human host and replicates efficiently in human alveolar epithelial cell line (A549) than other H9N2 sub-lineages. Due to the viral tropism, avian influenza viruses reach high titers when grown in chicken-origin cells (14-16). The efficient replication and infectivity of low pathogenic influenza viruses is achieved in the presence of supplemental trypsin or the type II transmembrane serine proteases (TMPRSS) (14, 17). In this study we provide cellular model to investigate the replication characteristics of H9N2 virus in either mammalian or avian-origin cells and compared their replication efficiencies in seven subsequent passages. We was examined the supernatants from each passage and determined if any changes occurred within the viral genome sequences.
2. Methods and Materials
2.1. Virus and Cell Cultures
A high-growth H9N2 virus belongs to G1 sub-lineage (18) was used for infecting the chicken embryo fibroblast (CEF) and A549 cells in this study. CEF cells were prepared from 10-day-old embryonated SPF eggs and maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and antibiotic solution. A549 cells (ATCC CCL-185TM) were cultivated in DMEM medium according to the instruction. Before the infection, expression of viral activating protease (VAP) and TMPRSS2 mRNAs in cell cultures were determined using the one-step RT-PCR mixture (Maxime RT-PCR premix; iNtRON Biotechnology, Korea).
Primers are shown in Table 1. Both proteases were expressed in A549, whereas TMPRSS2 was not detected in the CEF cells. Therefore, A549 cell culture was infected with H9N2 virus at multiplicity of infection (MOI) of 0.1 PFU/cell in 1 mL in DMEM containing 10% FBS, antibiotics solutions with and without supplemental trypsin and CEF cell was infected only in the presence of trypsin. Following adsorption for 1 hour at 37°C, the inoculum was removed and DMEM replaced. Up to 96 hours post-infection (hpi) and at 16-h intervals, culture supernatants were collected and stored at −70°C until used. The effect of cell passage on virus replication dynamics was determined by seven consecutive passages. In case, supernatant of each passage served as the virus seed for the next passage. The virus growth dynamics were evaluated by virus titration of cell culture supernatants at different hpi by tissue culture infection dose (TCID 50 ) assay.
| Gene | Forward Primer | Reverse Primer | Length, bp | Accession, No. |
|---|---|---|---|---|
| HA | CTCGAGCAAAAGCAGGGGAATTTCT | AAGCTTTTATATACAAATGTTGCACCT | 1761 | FJ794817 |
| NA | CTCGAGAGCAAAAGCAGGAGTAAAAATG | AAGCTTAGTAGAAACAAGGAGTTTTTT | 1431 | JX456183 |
| NS | CTCGAGAGCAAAAGCAGGGTGACAAAAAC | GGATCCAGTAGAAACAAGGGTGTTTTTA | 890 | JX308782 |
| VAP | TGCTGCTCATTGCATAAACC | GGGACTTCGAGCACTTTCAG | 330 | NM205022 |
| TMPRSS2 | ATCGACAAATGAGGGCAGAC | GTAGGCTGGGGACACTACCA | 480 | AF329454 |
| β-actin | TGCTGTGTTCCCATCTATCG | TTGGTGACAATACCGTGTTCA | 150 | L08165 |
The Primer Sequences Used for Amplification of the Studied Genes
2.2. TCID50 Assay
Aliquots of viral supernatants were 10-fold serially diluted with PBS, applied in each A549 and CEF cells/well of a 96-well plate, and incubated at 37°C for 1 hour. The inoculum was removed, and the cells were washed with PBS and supplied with DMEM containing FBS and trypsin. At seven consecutive passages, the TCID50 was determined based on the Reed-Muench method (19).
2.3. Antigenicity Test
To measure the antigenicity of viruses in each cell passage, H9N2 reference antisera (20) was used to measure antibody titers using the standard hemagglutinination inhibition (HI) assay (19).
2.4. Fusion Assay
The low-pH treatment was performed on H9N2 infected cells with the reaction buffer (DMEM, pH 5.0) for 5 minutes at 37°C. The buffer was removed and the cells incubated for 8 hours at 37°C in fresh neutral DMEM containing 5% FBS. Cells were fixed with ethanol and stained with Giemsa solution. Fusion activity was determined by counting the fused cells in an entire field.
2.5. Virus Genes Nucleotide Sequences
To investigate the effect of adaptation in CEF and A549 cells, full-length viral HA, NA, and NS genes were detected for passages 2, 4, 6 and 8. The primer sets listed in Table 1 were designed for amplification of the genes. The RT-PCR reaction consisted of 1 cycle at 42°C for 60 minutes and 95°C for 5 minutes and 30 cycles at 94°C for 30 seconds, 57.5°C for 45 seconds, and 72°C for 1 minute, followed by a final elongation step of 72°C for 10 minutes. The purified PCR products of viral genes at different passages were sequenced in both directions.
3. Results
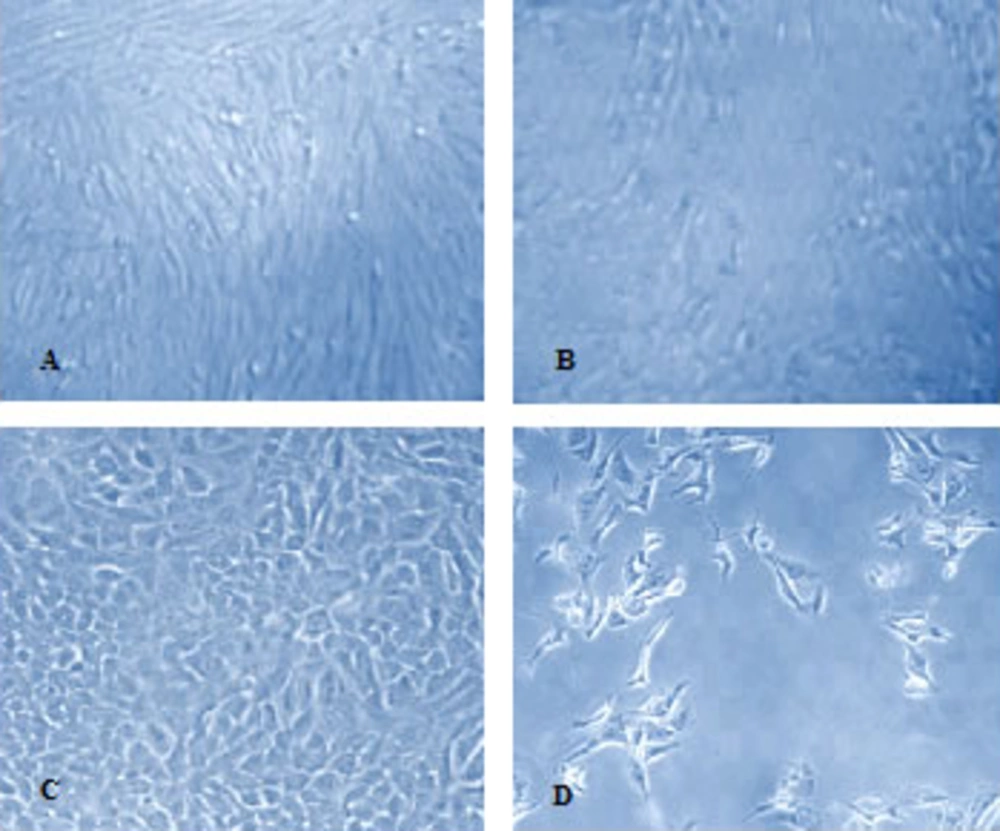
The H9N2 virus was well replicated in CEF cell and showed the cytopathic effect (CPE) in the presence of trypsin. The CPE was appeared in infected A549 cells with or without trypsin treatment Figure 1.
The infection of A549 cells in the absence of trypsin recognizes VAP and or TMPRSS2 as HA-processing proteases for the low pathogen subtype virus. The TCID 50 results showed a decrease in virus titer on the first passage which remained constant during the subsequent passages (Figure 2).
The CEF and A549 cells supplemented with trypsin were produced 3.6 and 3.8log10TCID50/mL, respectively which were less than the corresponding amount in chicken embryonated eggs (6.7). The H9N2 virus–infected A549 cell culture supplemented with trypsin exhibited the higher virus titer in shorter time compared to the trypsin-free infected cells. The HI results indicate the H9N2 virus in subsequent cell passages have similar antigenicity in both cell cultures. The average number of fused cell per field in A549 cells was not significantly different (46.42 in trypsin-treatment cell vs. 41.35 in non-treatment cell), confirming that the cell supported fusion and virus entry with or without trypsin treatment. The expression of the VAP and TMPRSS2 mRNAs in A549 cells, confirmed the role of cellular proteases in activation of the low pathogenic H9N2 virus.
Nucleotide sequences of the HA, NA, and NS genes of the virus passaged in the cell cultures were analyzed and compared with the parental virus. The comparative sequence analysis indicated several simultaneously nucleotide substitutions resulting in amino acid changes were occurred in NA of the virus replicated in A549 cells. Two fixed amino acid substitutions at positions G320 to A and G414 to A of NA were found up to the sixth passage. The amino acid sequences at the active sites of NA protein at positions 366 IRKDSRAG 373, 399 DSDNRSGY 406, and 431 PQE 433 were conserved. No amino acid changes were observed at the cleavage site, the receptor binding pocket and within the N-glycosylation sites of HA protein. All of them showed conservation of residues H183, L190, L226, Q227 and G228 in the receptor binding pocket and RSSR motif in cleavage site. All of the viruses possessed the typical avian ESEV motif in PDZ domain at the C-terminal end of NS1 protein. The substitution of amino acid D to E in position 92 which is required for high virulence was not shown.
4. Discussion
The replication of enveloped influenza viruses in host cells is a polygenic procedure depends on the host cell endocytic pathways for entry and transfer of viral genome as well as activation of host cell signaling (21, 22). The viruses replicate primarily in the epithelial cells of lung and intestines, then spread to the resident macrophages and recruited type II monocytes (23). A variety of cells have been evaluated to investigate the genetic basis of avian influenza viruse hosts. To gain a better understanding of how influenza viruses adapt to the new host, we passaged a high-growth H9N2 influenza strain in two different origin cells and determined replication efficiency of the virus and the molecular basis of adaptation.
The viral titers were decreased in CEF and A549 cells at first passage which remained constant up to the last passage. The final viral titer of 2.2 log10 TCID50/mL−1 was detected in trypsin-free A549 infected cells suggests that under this condition, virus replication was not prevented but clearly delayed. The peak of cell viral titer was accessed after 48 hours. This is in agreement with Sutejo and coworkers (24) suggested that H9N2 virus replicated less efficiently in A549 cell type. The appearance of CPE in A549 cell culture in the absence of trypsin correlates with the cleavage of HA protein. Böttcher et al. (12) have been reported that TMPRSS2 expressed in human airways is virus-activating protease that cleaves the monobasic cleavage motif of a H3N2 influenza virus. We show that the protease is able to activate the monobasic cleavage site of the H9N2 virus and support its infection in A549 cells. The A549 cells are permissive to the H9N2 virus growth, but fibroblastic cells that lack the enzymes, require additional trypsin for virus replication (14, 25).
The cell fusion was detected in infected A549 cells without supplemental trypsin, therefore; incomplete infection can be a reason for the lower virus titers. Addition of trypsin mediates fast propagation of influenza virus infection and leads to escape viruses from host defense mechanisms (26). The interferon (IFN) response is one of the main host defense mechanisms which expressed during influenza viruse infections. The outcome of IFN induction during influenza virus infection and its suppression by NS1 as well as its effect on virus replication indicate that the antiviral state inhibits virus replication at strain-specific level (27, 28). In human cells the replication of the avian influenza viruses is correlated with activation of types I and III IFN and cell-death signaling pathway (24).
A549 cells are IFN-competent and in the absence of trypsin the infection may result in a considerably stronger induction of IFN signaling and apoptosis, which significantly reduced the virus yields. However, the abilities of the cells to support the growth of avian influenza viruses are still debated because of possible change in receptor specificity in avian and the mammalian, susceptibility to higher concentrations of exogenous trypsin, etc. Results of this study demonstrated after seven serial passages of H9N2 virus in the cell cultures, the viral titer was not increased, which is indicating that the avian virus has not adapted to the human tissue culture; even if the viral antigenicity and sequencing patterns of the viral genes were stable.