1. Background
Klebsiella pneumoniae is responsible for up to 10% of all nosocomial infections (1, 2). The importance of the organism in hospital settings has been increasing due to the emergence and progressive spread of multidrug resistance; specifically the extended-spectrum β-lactamase (ESBL)-producing strains (3). More than 600 ESBL variants have been described and the majority of them belong to the SHV, TEM and CTX-M families (http://www.lahey.org/studies/webt.htm) (3). Horizontal gene transfer due to mobile genetic elements such as insertion sequences, transposons and conjugative plasmids, mediates intra and interspecies dissemination of not only the genes encoding ESBLs but also other antibiotic resistance determinants which are likely to form part of an antibiotic resistance integron (ARI) (3-5).
Three classes of ARIs (classes 1, 2, and 3) have been historically involved in multi-drug resistant (MDR) phenotypes and are identified based on their respective integrase genes (5). Various typing methods have been applied to understand transmission patterns of resistance genes and management of nosocomial infections (6). We have previously developed an optimized RAPD-PCR protocol for genotyping K. pneumoniae, comparable to PFGE (7). To understand the associations between phenotypic and genetic characteristics of multi-drug resistant pathogens can be useful for reliable detection of these bacteria in epidemiological studies. Some reports have suggested associations between ESBL production and resistance to several classes of antibiotics, as well as blaESBL with ARI genes carriage in K. pneumoniae (4, 8).
2. Objectives
In this study, the association between MDR phenotypes, prevalence of ARIs, blaESBL genes and RAPD profiles were investigated among ESBL-producing K. pneumoniae nosocomial isolates.
3. Materials and Methods
3.1. Bacterial Strains
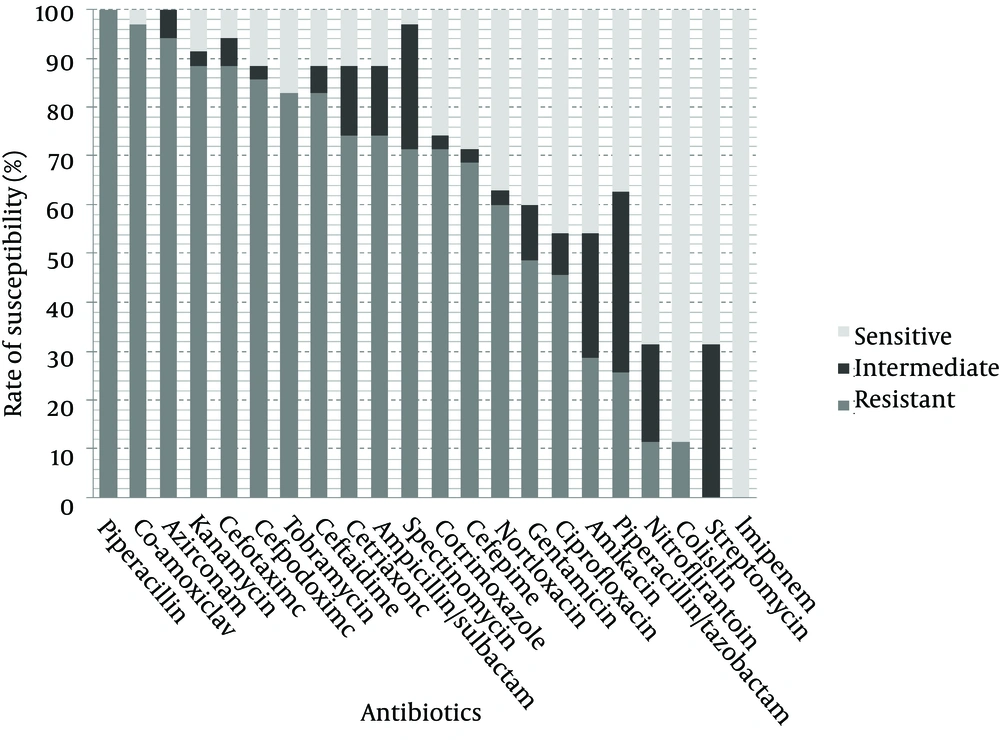
Thirty five ESBL-producing nosocomial isolates of K. pneumoniae were randomly selected from a collection previously described (9). Bacteria were isolated from hospitalized patients at different wards of Labbafinejad teaching hospital, Tehran, Iran, during March 2008 – March 2009; subjects consisted of 23 (65.7%) male patients and 12 (34.3%) females. These isolates were recovered from urine (n = 23; 65.7%), trachea (n = 4; 11.4%), wounds (n = 4; 11.4%), blood (n = 2; 5.7%), sputum (n = 1; 2.9%) and unknown sources (n =1; 2.9%). ESBL production was confirmed using the phenotypic confirmatory test and susceptibility of the isolates to 22 antimicrobial agents (Himedia, India) shown in Figure 1, was determined by the disc diffusion method according to the CLSI criteria (10).
3.2. Screening for Antibiotic Resistance Integrons
Genomic DNA was extracted from overnight grown bacteria using High Pure PCR template Prep kit for Genomic DNA extraction (Roche Diagnostics, Mannheim, Germany). PCR amplification of classes 1, 2 and 3 integrase genes was performed in 25 µL reaction mixtures containing 30 ng DNA template, 0.4 mM of each dNTP, 150 µM MgCl2, 0.2 U Super Taq DNA polymerase (CinnaGen, Tehran, Iran) and 1 pmol of each primer (FazaBiotech, Tehran, Iran) as follows: Int1F; CCTCCCGCACGATGATC, Int1R; TCCACGCATCGTCAGGC, Int2F; TTATTGCTGGGATTAGGC, Int2R; ACGGCTACCCTCTGTTATC, Int3F; AGTGGGTGGCGAATGAGTG, Int3R; TGTTCTTGTATCGGCAGGTG) (11).
Amplifications were performed in a Bioer TC25/H Thermal Cycler (Bioer Technology Ltd, Hangzhou, China) using the following program: initial denaturation at 95ºC for 5 minutes followed by 35 cycles of 1 minute at 94ºC, 1 minute at 60ºC and 1 minute at 72ºC with a final extension at 72ºC for 10 minutes. The amplified PCR products were resolved by electrophoresis in 1% agarose gels and visualized after staining with ethidium bromide.
3.3. Genetic Fingerprinting and Characterization of blaESBL Genes
Genetic profiles of the isolates by RAPD, confirmed by PFGE have been reported in our previous article (7). Presence of blaESBL genes (blaSHV, blaTEM and blaCTX-M) and the sequencing result for the isolates were also previously reported (9).
3.4. Statistical Analyses
To assess the strength and statistical significance of correlations between the studied variables including patient gender, type of specimen, antimicrobial susceptibility, MDR phenotypes (resistance to 6 or more antibiotics), carriage of ARIs, blaSHV, blaTEM and blaCTX-M genes and genotype grouping, and also measure the association between resistance to each of the aminoglycoside, quinolone and sulfonamide antibiotics, separate bivariate analyses were performed by use of the non-parametric Spearman's rank correlation test. To confirm the association between each pair of significantly correlated variables after factoring out the effect of other effective variables, partial correlation analyses were used. To interpret the results of correlation analyses, we considered correlation coefficients (r values) as well as the levels of significance (P values).
4. Results
The antibiotic susceptibility results are shown in Figure 1. As observed, all isolates were resistant to piperacillin followed by 97.1% resistance to co-amoxiclav, 94.3% to aztreonam, 88.6% to kanamycin and cefotaxime, 85.7% to cefpodoxime, 82.9% to tobramycin and ceftazidime, 74.3% to ceftriaxone and ampicillin/sulbactam, 71.4% to spectinomycin and cotrimoxazole, 68.6% to cefepime, 60% to norfloxacin, 48.6% to gentamicin, 45.7% to ciprofloxacin, 28.6% to amikacin, 25.7% to piperacillin/tazobactam and 11.4% to nitrofurantoin and colistin. All isolates were susceptible to imipenem. Streptomycin resistance was not observed but 31.4% of the isolates showed intermediate resistance. The most active antibiotic was imipenem followed by streptomycin, colistin and nitrofurantoin. Significant associations were observed between resistance to kanamycin, tobramycin, gentamicin, amikacin, norfloxacin, ciprofloxacin and cotrimoxazole (Table 1). Class 1 integrons were detected in 21 isolates (60%) and class 2 integrons in 3 isolates (8.57%). Two of the isolates carried both classes and none harbored class 3 integrons.
| Antibiotic classes | Antibiotic | Aminoglycosides | Quinolones | Sulfonamides | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KM a | TN a | GM a | AK a | SM a | NOR a | CIP a | TS a | ||||||||||
| r b | p c | r | p | r | p | r | p | r | p | r | p | r | p | r | p | ||
| Aminoglycosides | KM | -- d | -- | 0.789 | 0.1% | 0.412 | 5% | 0.363 | 5% | NS | NS | 0.456 | 1% | NS | NS | NS | NS |
| TN | 0.789 | 0.1% | -- | -- | 0.447 | 1% | 0.460 | 1% | NS | NS | 0.421 | 5% | NS | NS | NS | NS | |
| GM | 0.412 | 5% | 0.447 | 1% | -- | -- | 0.436 | 1% | NS | NS | NS | NS | 0.470 | 1% | NS | NS | |
| AK | 0.363 | 5% | 0.460 | 1% | 0.436 | 1% | -- | -- | NS | NS | NS | NS | NS | NS | NS | NS | |
| SM | NS | NS | NS | NS | NS | NS | NS | NS | -- | -- | NS | NS | NS | NS | NS | NS | |
| Quinolones | NOR | 0.456 | 1% | 0.421 | 5% | NS | NS | NS | NS | NS | NS | -- | -- | 0.504 | 1% | 0.410 | 5% |
| CIP | NS | NS | NS | NS | 0.470 | 1% | NS | NS | NS | NS | 0.504 | 1% | -- | -- | NS | NS | |
| Sulfonamides | TS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | 0.410 | 5% | NS | NS | -- | -- |
aAbbreviation: KM, kanamycin; TN, tobramycin; GM, gentamicin; AK, amikacin; SM, Streptomycin; NOR, norfloxacin; CIP, ciprofloxacin; TS, co-trimoxazole; NS, non-significant.
bCorrelation coefficients range between -1 (perfect negative relationship) and +1 (perfect positive relationship). A value of 0 indicates absence of any linear relationship.
c Level of significance.
dnot available.
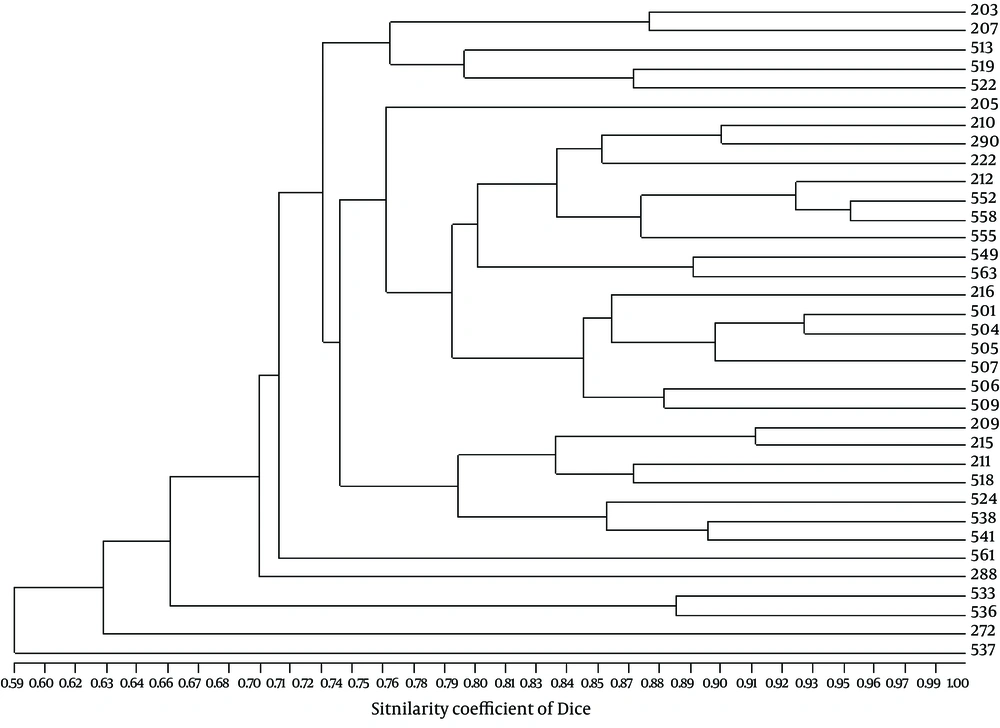
Genetic profiles of the isolates by RAPD (Figure 2) which were confirmed by PFGE, showed six major clusters (a-f) on a similarity level of 70%, and 21 different groups on a similarity level of 85% (7). Characterization of blaESBL genes from our previous work showed that 27 isolates (77.1%) harbored blaSHV genes including blaSHV-12, blaSHV-5 and blaSHV-11, 17 (48.6%) carried blaTEM genes characterized as blaTEM-1 by sequencing, 16 (45.71%) carried blaCTX-M-I which belonged to blaCTX-M-15 and 10 (28.57%) contained blaCTX-M-III characterized as blaCTX-M-8 (9).
Genotyping results were significantly correlated with carriage of ARIs (r = 0.700, P < 0.001; Spearman rank correlation test), blaSHV (r = 0.742, P < 0.001) and MDR phenotype (r = 0.560, P < 0.001). Significant association was also found between ARI carriage and MDR phenotype (r = 0.398, P < 0.05). Although at the 95% confidence level, no significant association was observed between ARIs with blaSHV, blaTEM and blaCTX-M among the isolates. A positive association was found between class 1 integrons with blaSHV-11, blaSHV-5 and blaSHV-12 at a lower confidence level (r = 0.298, P < 0.1) (Table 2). Results of partial correlation analyses were also confirmatory (data not shown). No correlation was observed between the patient gender or specimen source with any of the genetic variables.
aAbbreviations: r, correlation coefficient; NS, non-significant.
bcoefficients range between -1 (perfect negative relationship) and +1 (perfect positive relationship). A value of 0 indicates absence of any linear relationship.
5. Discussion
Infections due to ESBL-producing strains, have been most commonly reported regarding K. pneumoniae (3). ESBL encoding genes are usually located on plasmids which may also carry other antibiotic resistance determinants. Reports have suggested a close association between ESBL production and ciprofloxacin resistance in K. pneumoniae (8). Co-resistance with other classes of antibiotics such as fluoroquinolones, aminoglycosides, tetracyclines, chloramphenicol and sulfonamides are also widespread among ESBL producing strains (12). This may explain the significant associations found between resistance to aminoglycosides (kanamycin, tobramycin, gentamicin and amikacin) in this study. The same trend was observed for the association of resistance between norfloxacin with kanamycin, tobramycin, ciprofloxacin and cotrimoxazole. Similarly, resistance to ciprofloxacin and gentamicin were associated showing a relationship as a sign of co-carriage.
Bivariate correlation analyses followed by partial correlation analyses in order to distinguish between direct and indirect interactions, confirmed the results. Despite high heterogeneity observed among the isolates of this study, genotyping results were strongly correlated with carriage of ARIs and blaSHV genes. Although almost all K. pneumoniae isolates carry chromosomal non-ESBL blaSHV-1, nearly all ESBL encoding blaSHV genes found in K. pneumoniae are plasmid borne (13, 14). In this study, RAPD profiles were strongly correlated with the presence of blaSHV genes suggesting that plasmid mediated blaSHV-5 and blaSHV-12 (the two prevalent ESBL encoding blaSHV genes among our isolates) had some influence on RAPD patterns. Possible contribution of plasmid DNA to RAPD patterns was suggested in K. pneumoniae (15). However, Elaichouni et al. found no influence of plasmid DNA on the RAPD profiles in Escherichia coli and claimed that the amount of chromosomal DNA per cell in natural conditions inhibits observable plasmid amplification (16). The association of blaESBL genes with ARIs occurs when both form parts of complex integrons or are located on the same plasmid (4, 17).
We found a positive association between class 1 integrons and blaSHV-11, blaSHV-5 and blaSHV-12 at the confidence level of 90% (P < 0.1). Since genotyping results were highly correlated with the carriage of both ARIs and blaSHV, it could be concluded that ARIs and blaSHV genes are carried on the same plasmids, or blaSHV genes are located within ARIs at least among some of our isolates. Association between ARIs and blaSHV-5 as well as co-location of blaSHV-12 and a class 1 integron on the same plasmid have been reported (17, 18). However, other investigators have found a low rate of association between integrons and ESBL genes with the exception of blaCTX-M-9 (19).
Presence of plasmids that carry ESBL encoding genes as well as integron mediated antibiotic resistance has been reported among nosocomial isolates of K. pneumoniae (17, 19, 20). In most of these studies, ESBL encoding genes were located on plasmids but not within the integrons. Although most of the findings so far suggest contribution of integrons in the acquisition and transmission of resistance genes among bacteria, further investigations are needed to evaluate the involvement of other factors in transmission of linked resistance genes.