1. Background
Leishmaniasis is among the six most important tropical diseases while its different aspects are recommended as research topics by the World Health Organization (1, 2). Three forms of leishmaniasis including: Zoonotic Cutaneous Leishmaniasis (ZCL), Anthroponotic Cutaneous Leishmaniasis (ACL) and Visceral Leishmaniasis (VL) have been causing some health and medical problems related to humans and animals in Iran and its adjoining countries such as Iraq, Afganistan and Pakistan (3, 4). Cutaneous Leishmaniasis (CL) is considered as an important health problem in almost all countries of the Middle East including Iran (1, 2).
Cutaneous leishmaniasis is endemic in two forms ACL and ZCL, however ZCL is endemic in the Khuzestan province and caused by the zooflagellate parasite Leishmania major (5, 6). The main vector of this parasite in Iran, including Khuzestan is female Phlebotomus papatasi Scopoli (Diptera: Psychodidae). Tatera indica, the Indian gerbil, is the main reservoir host of L. major in the southwest of Iran (2, 5). Estimate shave shown an increase in the rate of ZCL cases in different regions of Khuzestan including western districts among the human population during the passed decade (7-9).
2. Objectives
One of these areas was Roffaye near the border with Iraq. Therefore, the present study was carried out to detect the nature of this parasite among trapped sandflies and rodent reservoir.
3. Materials and Methods
3.1. Study Area
Dasht-e-Azadegan County is located in the north-west of Ahvaz and has a border with Iraq. Latitude and longitude of this county are 31˚ 33' N and 48˚ 10' E, respectively. Altitude from sea level is 10 m. It includes 3 regions; central, Hoveyzeh and Boustan. Hoveyzeh region has 2 cities; Rofayyeh and Hoveyzeh. Rofayyeh (31˚ 55' N, 47˚ 40'), the area of this research study is situated 25 km from the west of Hoveyzeh and 50 km from the southwest of Susangerd city, the center of Dasht-e-Azadegan, between the Iran and Iraq border (Figure 1).
3.2. Sand fly and Rodent Collection and Identification
In this study sand flies were collected from indoors and outdoors using 150 sticky traps and the rodents using Sherman live traps from outdoor areas from May 2011-October 2011, in 3 attempts. All the traps were set at dusk and collected at dawn, the next day. All traps containing sandflies and rodents were transferred to the Dept. of Medical Entomology and Infectious and Tropical Diseases Research Center labs of Ahvaz Jundishapur University of Medical Sciences (AJUMS) to be identified. Rodent trapping and sticky traps for sand flies were performed in a rural region near the residential part of some villages. In these regions burrowing activities of rodents were seen. However, observations showed that some of residents had CL lesions. This suggests that this region may be an appropriate place for point of epidemiologic and ecologic conditions regarding CL. Therefore, by trapping 50 sand flies and 15 rodents we did the molecular experiments.
The external morphological characteristics in males including: head, claspers and terminal abdomen segments and internal morphological characteristics in females (spermatheca) were used to identify the species of sand flies (10). Next, they were identified at the species level using keys recommended by Lewis (11), Nadim and Javadian (12) and Nadim et al. (13).The rest of the abdominal segments in females were used to detect the L. major by Nested PCR application. The rodents were anesthetized using ether and the slide smears of their ear lesions were prepared using Geimsa staining. Their skull and tooth structures were used for their identification. The Iranian rodent key of Etemad was performed to identify the rodents (14).
3.3. Molecular Characterization
3.3.1. DNA Extraction
Smears were prepared from lesions. Rodents ear DNA was extracted by using High Pure PCR Template Purification Kit (Roche, Germany), according to the manufacturer’s instructions.DNA was extracted as described by Aransay et al. (15), for sand flies. Briefly, individual sand fly bodies were homogenized with a sealed Pasteur pipette in 1.5-ml tubes. One hundred fifty microliters of extraction buffer (1% sodium dodecyl sulfate [SDS]–25 mMNaCl–25 mM EDTA) was added, and samples were placed at 65°C for 30 min. Following the addition of 100 ml of 3 M potassium acetate (pH 7.2), the homogenates were incubated on ice for 30 min and then centrifuged for 15 min at 13,000 3 g. Supernatants were recovered, and DNA was precipitated with the addition of 600 ml of 100% ethanol. DNA pellets were re-suspended in 50 ml of 0.53 Tris-EDTA (TE) (pH 8.0). Five-microliter portions of these DNA extracts were used for PCR amplification.
3.3.2. Amplification of Kinetoplastic MinicircleDNA From Sand Flies and Rodents
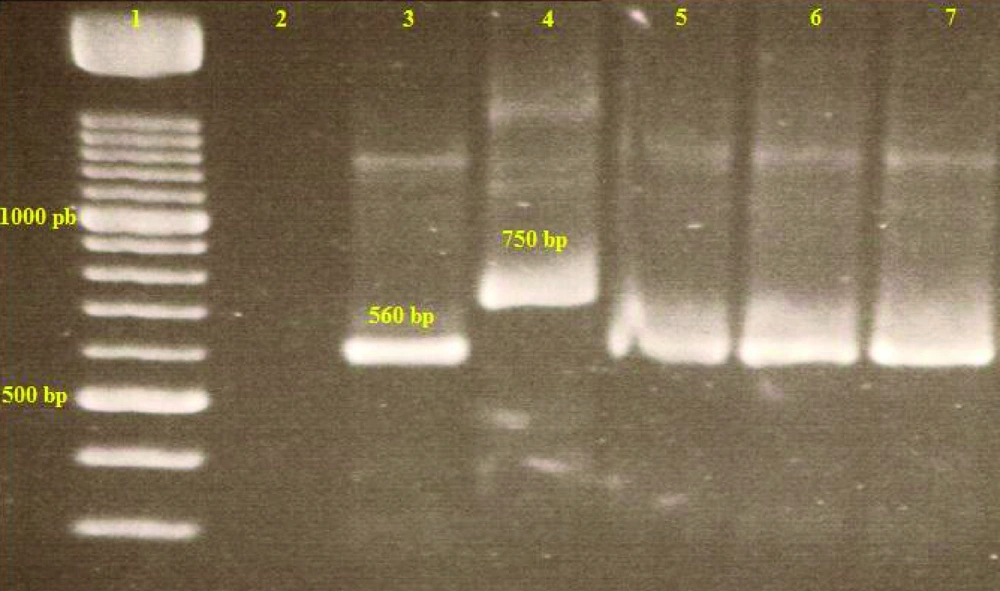
For amplification of variable minicircles of Leishmania kDNA, Nested – PCR was performed according to previous studies as follows: in the first stage two external primers CSB2XF (CGAGTAGCAGAAACTCCCGTTCA) and CSB1XF (ATTTTTCGCGATTTTCGCAGAA CG) and in the second step, two internal specific primers 13Z (ACTGGGGGTTGGTGTAAAATAG) and LiR (TCGCAGAACGCCCCT) were used (15, 16). Ready to use Bioneer, Accupower PCR premix, 96 tubes, 0.2ml and 20µlrxn (Bioneer Co., Korea) were used for the PCR reaction. DNA was amplified using thermal cycler (Eppendorf AG 22331, Hamburg, Germany) under the following conditions: 5 min at 94°C followed by 35 cycles of 30 sec at 94°C, 60 sec at 55°C, 90 sec at 72°C and a final elongation at 72°C for 10 min. For each sample, one positive control and one negative control were included. Two microliters of the first-round product was used as a template for the second round in a total volume of 20 μl under the same conditions as those for the first round, except with primers LiR and 13Z.The PCR products were visualized by 1.5% agarose gel electrophoresis, using a 100- bp DNA ladder marker at 260 nm. Results were compared with standard band markers of L. major (MHOM/IR/75/ER) providing fragment of 560 bp (17).
4. Results
Totally 50 sandflies were trapped during this study. All of them were identified as P. papatasi, 27 females and 23 males, from outdoor places (15 females and 8 males) and indoor places (12 females and 15 males). Totally 15 rodents including 12 T. indica (Indian gerbil) and 3 Allactaga sp. were trapped in the current study. The Iranian rodent keys recognized 12 of the trapped rodents as T. indica with the following character: each of the upper incisors has one longitudinal groove in the anterior surface. The upper molars have been facilitated with fragmented grooves. The length of occipitonasal was longer than 43 mm. The Zygomatic plate was extended forward. The tail of gerbils were darker in dorsal and ventral and lighter in the lateral sections (14).
Results of the second stage of the nested PCR technique showed that 2out of the 27 female P. papatasi (7.4%) and1smear (ear lesion) from 12 of the T. indica (8.3%) was positive to parasite due L. major (bands of 560bp) (Figure 2). The trapped Alactaga sp. Rodents were not infected with L. major . The positive smears of sandflies were made from indoor sandfly specimens.
5. Discussion
A ZCL control program is based on promoting our knowledge regarding the parasite’s reservoir host(s) and vectors (18). However, ZCL is endemic in Khuzestan but the majority of carried out studies have insisted on L. major of human specimens. Less studies have been performed on L. major of sandfly vectors and rodent reservoir host specimens in Khuzestan province and they belong to many years ago, when no molecular methods were used (2). P. papatasi has been confirmed as the main vector of L. major in Shoush area, north west of Khuzestan using classical entomology and parasitological methods (19).
There is a similar trend for T. indicaas reservoir of L. major in Khuzestan. All of the recent molecular Leishmania studies in Khuzestan are also related to human specimens. The current study is the first molecular report which describes naturally infected P. papatasi and T. Indica with L. major at the same time in the Roffaye region in the west of Khuzestan. The results of this study showed that a Nested-PCR as a sensitive and high-resolution method could detect L. major in both sand fly insects and gerbil rodents as vectors and reservoirs of the protozoan, respectively, from a region with a high rate of ZCL among the rural residents.
Results of the current study are consistent with the results of Azizi et al. (20) and Rassi et al. (21) regarding the same bands of L. major (560 bp) in P. salehi and P. papatasi, respectively (20, 21). However, same bands have been confirmed in human specimens of L. major in Khuzestan (9). The rate of L. major infection in the current study was 6.9% for female P. papatasi. This rate has been reported as 5.5% in P. salehi from south of Iran (21). T. indica has been confirmed as the main reservoir of ZCL in the south west of Iran including Shoush of Khuzestan using classical parasitological methods (inoculation of scrapings from the ear of rodents in white mice and observing amastigotes in the mice) (5).
The rate of infected T. indica was reported as 12.5%. In another study which was carried out in Ilam, T. indica was reported as the main reservoir of ZCL in Mehran with a rate of 5.5% (5). Results of the current study in terms of trapped T. indica showed that 8.3% of the rodents were infected with L. major as the first report in Khuzestan by a molecular method. This rate was recorded as 11.8% among trapped Gerbillusnanus, using PCR method (22) and 3.7% of trapped T. indica using classic parasitological methods in south-east of Iran (23).
According to the results of the current study, where molecular methods detected L. major in both P. papatasi and T. indica from same area and molecular reports regarding L. major in CL human specimens of different parts of west Khuzestan and similar results from classical parasitological methods, it can be concluded that P. papatasi and T. indica should be considered as a vector and reservoir of ZCL in Roffayeh. Consequently, this area could be considered as the focus of ZCL, which needs to be treated by control methods of ZCL removal.