1. Background
Staphylococcus aureus has remained a primary pathogen of nosocomial and community-acquired infections worldwide. Infection of this bacterium causes disease in different body organs and easily spreads in the hospital. The emergence and spread of methicillin-resistant and Vancomycine- intermediate S.aureus (VISA) isolates have increased their importance. Understanding the epidemiology and distribution of this bacterium in the community as well as hospitals is necessary to determine its source, to restrict its spread especially in hospital settings. Finding a suitable marker for this purpose has been considered for many years and different methods have been used. Phage typing is one of the oldest methods, which has been declined in the recent years because of its technical problems and the nature of its usage, and instead, molecular methods on the basis of coagulase gene typing (coa typing), agr gene and others have been included. Polymorphism of the gene encoding protein A (spa) is one of the methods used in several studies (1).
Protein A is one of the main cell wall proteins considered as the virulence factor of S. aureus (2). This protein is unique in this bacterium, as it can act as IgG binding site, except for IgG3. Although each protein A contains five IgG binding sites (A, B, C, D, E), some strains contain four (3). The gene encoding this protein (spa) is different in length in various strains of the bacterium, between 1150 to 1500 base pairs (bp). Another reason for the spa gene length difference is its X region diversity; there is a repeated unit composed of a 24-nucleotide region on the X region of the spa gene, repeated 2-16 times. These factors can cause major polymorphisms, used as the base of S. aureus classification by a high differentiation power (4, 5). Due to the high precision and discriminatory power of this method as well as wide variety of the spa types, today, several online sites are designed to identify the S. aureus type, such as NCBI and spa Ridom type.
However, one of the problems in these methods is need for sequencing, which is expensive. In addition, in this typing method, only X region of the spa gene is considered, and other changes are forgotten, especially in the antibody binding site. Therefore, in this study, we sought to replace the X sequencing using PCR-RFLP (polymerase chain reaction restriction fragment-length polymorphism) method for the spa typing (6).
PCR-RFLP of spa for tracking and typing methicillin-resistant S. aureus (MRSA) was proposed in 2005 by Mitani (7). Different Enzymes, especially HaeII, have been used for this purpose; but in this study, we digested the spa gene product with Bsp143I enzyme. The OLIGO software version 5 was applied to detect the restriction sites of this enzyme on the spa gene. The restriction sites are outside of the X region and digest the GATC sequence. In wild type isolates, there are at least three restriction sites and we can find four bands in electrophoresis, according to the standard S. aureus strain 8325.
2. Objectives
This study was designed for S. aureus typing based on PCR-RFLP of the spa encoding gene, using the Bsp143I enzyme.
3. Patients and Methods
3.1. S. aureus Isolation
This study was carried out on 128 isolated S. aureus samples collected from nose of 53 (41.4%) healthcare workers as healthy carriers as well as 75 (58.6%) from patients referred either to the teaching hospital or private medical diagnostic laboratory in Gorgan, north of Iran. Clinical samples were obtained from 24, 16, 17 and 18 cases of urine, wound, blood and other specimens (including synovial fluid, sputum and throat culture), respectively. Purification and determination of S. aureus were performed by biochemical methods, and its identification based on amplification of the glutamate synthetase gene was confirmed which was performed by PCR. Specific primers of mecA were applied to assess the MRSA strains; it was found that 45 (35.2%) and 83 (64.8%) of S. aureus isolates were MRSA and methicillin-sensitive S. aureus (MSSA), respectively (8).
3.2. DNA Extraction and PCR of spa
DNA extraction and spa gene amplification were carried out according to our previous study (8) which can be explained briefly as follows: DNA was extracted using lysostaphin and phenol-chloroform, and specific primers (4) (mentioned below) were used for PCR with product lengths varying between 1150-1500 bp;
spa1) 5′-ATCTGGTGGCGTAACACCTG-3′
spa2) 5′-CGCTGCACCTAACGCTAATG-3′
3.3. PCR RFLP
The Bsp143I enzyme (Fermentas, Germany) was used to digest the final PCR product of the spa gene. The procedures were according to the instructions of Fermentas Co.: 10 μL (~ 0.1-0.5 μg of DNA) of PCR reaction mixture was added to 18 μL of nuclease-free water, 2 μL of 10X buffer x Bsp143I buffer, and 2 μL of Bsp143I. The mixture was mixed gently and spin down for a few seconds and incubated at 37°C for four hours. Afterwards, Bsp143I was inactivated by incubation at 65°C for 20 minutes. The OLIGO software version 5 was also applied to detect the restriction sites on the gene. The restriction sites of this enzyme are outside the X region and it recognizes the “GATC” sequence. The final products after Bsp143I digestion were analyzed by electrophoresis on 1.5% gel. Finally, they were compared with S. aureus strain 8325-4 on NCBI.
3.4. Statistical Analysis
Statistical comparison of the results was performed by χ2 test and P < 0.05 was considered significant.
4. Results
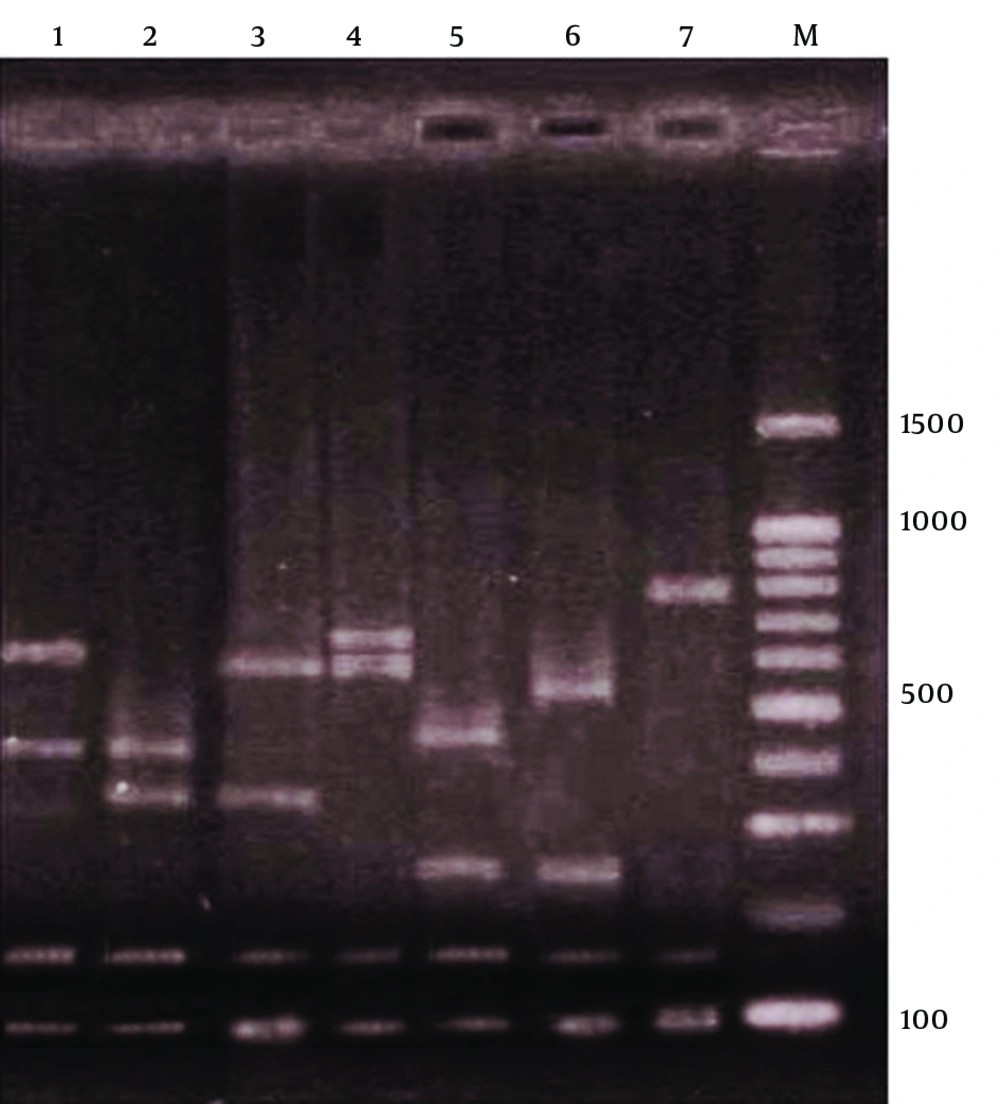
After PCR amplification, digestion of the spa gene by Bsp143I revealed seven types. Electrophoresis of the spa gene digestion showed four bands in six of the types, but in the seventh type, three bands were produced. Bands of 97 and 150 bp were present in all the seven types, but the sizes of other bands varied (Figure 1).
First and second spa gene types with 50 (39.06%) and 37 (28.9%) cases respectively, were the most common types, but only one S. aureus isolate belonged to the fifth to seventh spa types. The frequencies of the first to fourth spa types were as follows; type 1: 39.06%, 2: 28.9%, 3: 16.4%, and 4: 13.3%. Table 1 shows the distribution of seven spa types in MRSA and MSSA S. aureus samples, showing a statistically significant difference (P < 0.001). While type 1 (45.8%) was the most common among the MSSA isolates, type 2 (53.3%) was the most prevalent among MRSA isolates. On the other hand, spa type 3 was significantly more prevalent in MRSA than MSSA (Table 1). Table 2 shows the distribution of the seven spa types among S. aureus samples isolated from patients and healthy carriers. spa types 1 and 2 and spa types 1, 3 were predominant in S. aureus samples isolated from patients and healthy carrier, respectively. Difference in distribution of the spa types in the two groups was statistically significant (P = 0.003). Table 3 shows the frequency of all spa types in S. aureus samples isolated from the patients. While types 1, 2 and 4 were found in all the clinical samples, type 3 was only seen in the isolates from skin and throat culture.
| Type | 1 | 2 | 3 | 4 | 5 | 6 | 7 | Total |
|---|---|---|---|---|---|---|---|---|
| Carrier b | 17 (32.1) | 11 (20.8) | 17 (32.1) | 7 (13.2) | 0 | 0 | 1 (1.9) | 53 (100.0) |
| Patient | 33 (44.0) | 26 (34.7) | 4 (5.3) | 10 (13.3) | 1 (1.3) | 1 (1.3) | 0 | 75 (100.0) |
| Total | 50 (39) | 37 (28.9) | 21 (16.4) | 17 (13.3) | 1 (0.8) | 1 (0.8) | 1 (0.8) | 128 (100) |
a Data are presented in No. (%).
b P value of the spa types between carriers and patients was = 0.003.
| Type | 1 | 2 | 3 | 4 | 5 | 6 | 7 | Total |
|---|---|---|---|---|---|---|---|---|
| MSSA c | 38 (45.8) | 13 (15.7) | 20 (24.1) | 9 (10.8) | 1 (1.2) | 1 (1.2) | 1 (1.2) | 83 (100.0) |
| MRSA | 12 (26.7) | 24 (53.3) | 1 (2.2) | 8 (17.8) | 0 | 0 | 0 | 45 (100.0) |
| Total | 50 (39) | 37 (28.9) | 21 (16.4) | 17 (13.3) | 1 (0.8) | 1 (0.8) | 1 (0.8) | 128 (100) |
a Abbreviations: MRSA, methicillin-resistant S. aureus; MSSA, methicillin-sensitive S. aureus.
b Data are presented in No. (%).
c P value of the spa types between MRSA and MSSA was < 0.001.
| Type | Urine | Wound | Blood | Others b | Total |
|---|---|---|---|---|---|
| 1 | 12 (36.3) | 4 (12.1) | 6 (18.1) | 11 (33.3) | 33 (100) |
| 2 | 6 (23.1) | 7 (26.9) | 9 (34.6) | 4 (15.3) | 26 (100) |
| 3 | 0 | 3 (75) | 0 | 1 (25) | 4 (100) |
| 4 | 5 (50) | 1 (10) | 2(20) | 2 (20) | 10 (100) |
| 5 | 0 | 1 (100) | 0 | 0 | 1 (100) |
| 6 | 1 (100) | 0 | 0 | 0 | 1 (100) |
| 7 | 0 | 0 | 0 | 0 | 0 |
| Total | 24 (32) | 16 (12.5) | 17 (12.9) | 18 (13.6) | 75 (100) |
a Data are presented in No. (%).
bS. aureus isolated from other samples such as: synovial fluid, CSF, sputum, throat culture, and stool culture.
5. Discussion
Typing of pathogenic bacteria is an important process in epidemiological studies and control of nosocomial infections. spa typing and PCR-RFLP in addition to pulsed field gel electrophoresis (PFGE) are among the efficient methods for differentiating S. aureus, particularly MRSA strains (9). In this study, S. aureus isolates were classified by PCR-RFLP of the spa gene with Bsp143I enzyme, which classified in seven types.
The spa gene lengths in the isolated cases varied from 1150 to 1500 bp, which were slightly longer than the data reported from a study in India (1150-1420 bp). The maximum repetition of the X-region in our samples was 12 times, but the same item in a study from India was 13 (4). This discrepancy is probably due to differences in the IgG binding site of the spa gene, which is outside of the X region. Bsp143I is an endonuclease enzyme that digests the GATC sequence in DNA, which is used in several studies (10, 11). However, this is the first time this enzyme is applied for digestion of the spa gene for S. aureus typing. Using this method, we were able to divide the S. aureus isolates to seven types. In a similar study in India on 149 isolates of S. aureus, PCR product of the spa gene was digested with HaeII enzyme and five types were identified. In other studies using HaeII enzyme in Germany and Iran, 14 and seven types were diagnosed, respectively (4, 12).
These findings imply that efficacy of the Bsp143I enzyme in digesting the S. aureus spa gene is similar to HaeII enzyme. In accordance with a previous research, we found that variation of the spa types among MSSA is more than MRSA isolates, insomuch all the seven spa types were present in MSSA isolates, but only four (types1-4) were seen in MRSA. Fenner et al. diagnosed 65 and 42 spa types among 101 MSSA and 200 MRSA isolates, respectively (13). Strommenger et al. demonstrated that the types variations among 283 MSSA isolates (128 types) was more than the variations among 1176 MRSA isolates (121 types) (14). Methicillin resistance is a relatively new phenomenon caused by acquiring the mecA gene by S. aureus; thus, many strains of S. aureus may not still have enough time to acquire the resistance genes. On the other hand, believing that only some S. aureus strains have the ability to obtain the mecA gene, just certain types of S. aureus can be MRSA and diversity of the spa types is less than MSSA (8).
Variety of the S. aureus strains spa types isolated in this study among patients and healthy carriers was statistically significant. Although distribution of the type 1 spa gene was similar in both patients and healthy carriers, the frequency of type 3 among carriers was significantly more.
This probably means fewer virulence of type 3 spa. Although it easily colonized in the noses of health care workers, it was not important in the pathogenesis. Further studies are recommended to identify its frequency and possible reasons for its decreased pathogenicity.
Our results showed that the Bsp143I enzyme is a useful tool for digestion of the spa gene and typing of S. aureus. The spa types variations among MSSA were more than MRSA isolates.