1. Background
Staphylococcus aureus has long been considered as a major pathogen of hospital infections. These bacteria are one of the most commonly diagnosed causes of skin and soft-tissue infections (such as furunculosis, impetigo, wounds, burns and abscess), as well as systemic infections (such as pneumonia, Respiratory Tract Infections (RTIs) and Urinary Tract Infections (UTIs)) (1-5). One of the best therapeutic choices for treatment of cases of S. aureus is by administration of methicillin yet the majority of recent studies have shown that the numbers of methicillin resistant Staphylococcus aureus (MRSA) strains have increased (6-8). Approved research revealed that 45 to 50% of S. aureus strains of human clinical infections were methicillin resistant (9, 10).
The gene for methicillin resistance, mecA, is carried on a 21- to 67-kb element, the Staphylococcal Chromosomal Cassette mec (SCCmec), which integrates at a conserved location in the S. aureus genome (11, 12). Staphylococcal Chromosomal Cassette mec genetic elements are characterized by the presence of two essential genetic markers (mec and ccr gene complexes). These elements are classically classified into types I, II, III, IV and V, according to the nature of the mec and ccr genes (12, 13). Type IV of the SCCmec genetic element is divided to IVa, IVb, IVc and IVd alleles (11-13). Study of the distribution of the SCCmec element of S. aureus is essential for molecular typing of MRSA strains.
Accordingly, S. aureus is considered as one of the most resistant bacteria in cases of hospital infections (4, 14). Various investigations have shown that antibiotic resistance genes are the most effective factors for causing antibiotic resistance in S. aureus. The most commonly detected antibiotic resistance genes in S. aureus strains of human clinical infections are mecA (methicillin), linA (lincosamides), msrA and msrB (macrolides), vatA, vatB and vatC (acetyl transferase genes and streptogramin A), ermA, ermB and ermC (macrolide– lincosamide–streptogramin B) and tetK and tetM (tetracycline), and aacA-D (aminoglycosides) (15). The frequent recovery of staphylococcal isolates that produce leukocidal toxins from patients with UTIs, RTIs, soft tissue and skin infections, suggests that the Panton-Valentine Leukocidin (PVL) is a virulence marker with a significant role in the pathogenicity of diseases (1, 16). Panton-Valentine Leukocidin has also been associated with severe diseases including skin and soft-tissue and systemic infections (1, 16).
2. Objectives
Based on the scarce data in the field of S. aureus and its antibiotic resistance properties in pediatric patients, the present research was carried out in order to study the prevalence of MRSA in various types of clinical infections of pediatric patients as well as to determine the distribution of PVL gene, SCCmec type, antibiotic resistance genes and antibiotic resistance pattern of bacterial isolates.
3. Patients and Methods
3.1. Samples Collection and Methicillin Resistant Staphylococcus aureus Identification
Overall, 255 clinical samples from various types of infections including blood (n = 40), UTIs (n = 60), respiratory tract infections (n = 55), superficial and post-surgical wounds (n = 50) and burn infections (n = 50) were collected from hospitalized pediatrics of four major provinces of Iran (Tehran, Isfahan, Mashhad and Shiraz). All samples were immediately transferred to the laboratory at 4°C in a cooler with ice packs.
Twenty-five microliters of each sample was inoculated on homogenized Mueller–Hinton Broth (MHB, Merck, Germany) supplemented with 6.5% NaCl. The suspension was incubated for 16 – 20 hours at 37°C. One milliliter of the enriched MHB media was added to 9 mL of phenol red mannitol broth containing ceftizoxime (5 μg/mL) and aztreonam (75 μg/mL) (PHMB), and incubated for 16 – 20 hours at 37°C. The surface of the selective isolation medium (MRSA ID) (MRSA ID is a new chromogenic medium for identification of MRSA in human specimens and was supplied as prepoured culture plates from bioMérieux) was inoculated with a sterile loop. The plates were incubated for 24 hours at 37°C (when the colonies were difficult to identify the incubation was protracted for another 24 hours). Typical green colonies were primary known as MRSA. Five selected typical colonies per plate were subcultured on Tryptone Soya Agar (TSA, Merck, Germany). Typical colonies were tested with the Staphytect Plus test (Oxoid); a latex agglutination test for the detection of clumping factor, Protein A and certain polysaccharides found in MRSA.
3.2. DNA Extraction and Polymerase Chain Reaction Confirmation of Methicillin Resistant Staphylococcus aureus Strains
Bacterial strains were grown overnight in TSA at 37°C. The grown bacteria were centrifuged at 800 μg for 10 minutes and the supernatant was discarded. The sediments were resuspended in 5 mL Phosphate Buffered Saline (PBS), shaken for 30 minutes and then re-centrifuged at 800 × g. The washed bacterial cells were transferred to 1.5mL microtubes and 10 μL achromopeptidase (10000 u/mL) was added and incubated at 55°C for 30 minutes. Next, 10% Sodium Dodecyl Sulfate (SDS) plus 5 μL (10 mg/mL) of proteinase K were added and the tubes were incubated in a water bath at 37°C for one hour. The resulting suspension was extracted twice with phenol/chloroform and once with chloroform. Each step of the extraction was achieved using a sterile microtube and centrifuged at 12500 × g. The bacterial DNA was recovered from the supernatant by precipitated DNA in pure ethanol, which was kept overnight in a -20°C freezer. Precipitated DNA was washed with 70% ethanol to separate any protein contamination and then resuspended in 40 μL Tris-EDTA (TE) buffer. The suspension was used as a pure DNA.
The presence of MRSA was confirmed using PCR amplification of femA and mecA genes. The DNA samples, which were simultaneously positive for femA and mecA genes, were also confirmed to be MRSA. For this purpose the PCR method, which was introduced by Jonas et al., (2002) (17) was used. The PCR reactions were performed in a total volume of 25 μL, including 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 200 μM dNTPs each (Fermentas, Germany), 2.5 μL PCR buffer (10X), 25 pmoL of each primer (MecA1 (5'-GTAGAAATGACTGAACGTCCGATAA-3') and MecA2 (5'-CCAATTCCACATTGTTTCGGTCTAA-3') (310 bp) and FemB1 (5'-TTACAGAGTTAACTGTTACC-3') (651 bp)), 1.5 U of Taq DNA polymerase (Fermentas, Germany) and 5 μL (40-260 ng/μL) of the extracted DNA template of the MRSA isolates. The DNA thermo-cycler (Eppendorf Mastercycler 5330, Eppendorf-Nethel-Hinz GmbH, Hamburg, Germany) was used in all PCR reactions for DNA amplifications. The PCR cycling conditions were as follows: initial denaturation at 94°C for four minutes, followed by 30 cycles of 45 seconds at 94°C, 45 seconds at 50°C, and 60 seconds at 72°C, with a final extension step at 72°C for two minutes.
3.3. Antimicrobial Susceptibility Test
The pattern of antimicrobial resistance was studied using the simple disk diffusion technique. The Mueller–Hinton agar (Merck, Germany) medium was used for this purpose. Antibiotic resistance of the MRSA strains against 18 commonly used antibiotics was determined using the instructions provided by the Clinical and Laboratory Standards Institute guidelines (18). Susceptibility of MRSA isolates was tested against ampicillin (10 µg/disk), gentamycin (10 µg/disk), lincomycin (2 µg/disk), cephalothin (30 µg/disk), imipenem (30 µg/disk), tetracycline (30 µg/disk), vancomycin (5 µg/disk), ciprofloxacin (5 µg/disk), norfloxacin (30 µg/disk), cotrimoxazole (30 µg/disk), clindamycin (2 µg/disk), trimethoprim-sulfamethoxazole (25 μg/disk), penicillin (10 µg/disk), oxacillin (1 µg/disk), erythromycin (15 µg/disk), azithromycin (15 µg/disk), ceftriaxone (30 µg/disk) and cefixime (5 µg/disk) antibiotic agents (Oxoid, UK). The plates containing the discs were allowed to stand for at least 30 minutes before incubation at 35°C for 24 hours. The diameter of the zone of inhibition produced by each antibiotic disc was measured and interpreted using the CLSI zone diameter interpretative standards (18). Staphylococcus aureus ATCC 25923 and Escherichia coli (E. coli) ATCC 25922 were used as quality control organisms in antimicrobial susceptibility determination.
3.4. Detection of Antibiotic Resistance Genes
Presence of antibiotic resistance genes in the MRSA isolates was analyzed using the method described previously (19, 20). Oligonucleotide primers and the PCR program used for amplification of antibiotic resistance genes are shown in Table 1.
| Target Gene | Primer Sequence (5'-3') | PCR Product, bp |
|---|---|---|
| AacA-D a | F: TAATCCAAGAGCAATAAGGGC | 227 |
| R: GCCACACTATCATAACCACTA | ||
| ermA a | F: AAGCGGTAAACCCCTCTGA | 190 |
| R: TTCGCAAATCCCTTCTCAAC | ||
| ermC a | F: AATCGTCAATTCCTGCATGT | 299 |
| R: TAATCGTGGAATACGGGTTTG | ||
| tetK a | F: GTAGCGACAATAGGTAATAGT | 360 |
| R: GTAGTGACAATAAACCTCCTA | ||
| vatC a | F: AAAATCGATGGTAAAGGTTGGC | 467 |
| R: AGTTCTGCAGTACCGGATTTGC | ||
| tetM b | F: AGTGGAGCGATTACAGAA | 158 |
| R: CATATGTCCTGGCGTGTCTA | ||
| vatA b | F: TGGTCCCGGAACAACATTTAT | 268 |
| R: TCCACCGACAATAGAATAGGG | ||
| msrA c | F: GGCACAATAAGAGTGTTTAAAGG | 940 |
| R: AAGTTATATCATGAATAGATTGTCCTGTT | ||
| msrB c | F: TATGATATCCATAATAATTATCCAATC | 595 |
| R: AAGTTATATCATGAATAGATTGTCCTGTT | ||
| vatB d | F: GCTGCGAATTCAGTTGTTACA | 136 |
| R: CTGACCAATCCCACCATTTTA | ||
| linA e | F: GGTGGCTGGGGGGTAGATGTATTAACTGG | 323 |
| R: GCTTCTTTTGAAATACATGGTATTTTTCGA |
Oligonucleotide Primers and Polymerase Chain Reaction Conditions for Detection of Antibiotic Resistance Genes in Methicillin Resistant Staphylococcus aureus Strains Isolated From Various Types of Clinical Infections in Pediatric Patients
3.5. Detection of Staphylococcal Chromosomal Cassette mec Types of The Methicillin Resistant Staphylococcus aureus Isolates
Table 2 shows the list of primers used for detection of SCCmec types in the MRSA isolates of various types of clinical infections (21). The multiplex PCR reactions were performed in a total volume of 50 μL, including 5 μL of 10 × PCR buffer (75 mM Tris-HCl, pH 9.0, 2 mM MgCl2, 50 mM KCl, 20 mM (NH4) 2SO4), 1 μL dNTPs (40 mM), 1 μL (50 pmol) of the forward and reverse primers, 5 μL of the extracted DNA template and 1 μL of 1U Taq DNA polymerase. The DNA thermo-cycler (Eppendorf Mastercycler 5330, Eppendorf-Nethel-Hinz GmbH, Hamburg, Germany) was used in all PCR reactions for DNA amplifications. The thermal cycler was adjusted as follows: 94°C for five minutes, followed by 10 cycles of 94°C for 45 seconds, 65°C for 45 seconds, and 72°C for 1.5 minutes, and 25 cycles of 94°C for 45 seconds, 55°C for 45 seconds, and 72°C for 1.5 minutes, followed by final extension at 72°C for 10 minutes and followed by a hold at 4°C. Fifteen microliters of PCR products in all reactions were resolved on a 2% agarose gel containing 0.5 mg/mL of ethidium bromide in Tris–borate–EDTA buffer at 90 V for one hour, using suitable molecular weight markers. The products were examined under ultraviolet illumination.
| Target Genes | Primer Sequence (5'-3') | Size of Product, bp |
|---|---|---|
| SCCmec I | F: GCTTTAAAGAGTGTCGTTACAGG | 613 |
| R: GTTCTCTCATAGTATGACGTCC | ||
| SCCmec II | F: CGTTGAAGATGATGAAGCG | 398 |
| R: CGAAATCAATGGTTAATGGACC | ||
| SCCmec III | F: CCATATTGTGTACGATGCG | 280 |
| R: CCTTAGTTGTCGTAACAGATCG | ||
| SCCmecIVa | F: GCCTTATTCGAAGAAACCG | 776 |
| R: CTACTCTTCTGAAAAGCGTCG | ||
| SCCmecIVb | F: TCTGGAATTACTTCAGCTGC | 493 |
| R: AAACAATATTGCTCTCCCTC | ||
| SCCmecIVc | F: ACAATATTTGTATTATCGGAGAGC | 200 |
| R: TTGGTATGAGGTATTGCTGG | ||
| SCCmecIVd | F: CTCAAAATACGGACCCCAATACA | 881 |
| R: TGCTCCAGTAATTGCTAAAG | ||
| SCCmec V | F: GAACATTGTTACTTAAATGAGCG | 325 |
| R: TGAAAGTTGTACCCTTGACACC |
Oligonucleotide Primers Used for the Detection of Staphylococcal Chromosomal Cassette mec types of the Methicillin Resistant Staphylococcus aureus Strains Isolated From Various Types of Clinical Infections in Pediatric Patients
3.6. Detection of Panton-Valentine Leukocidin Genes of Methicillin Resistant Staphylococcus aureus
Polymerase Chain Reactions (22) were performed in a final volume of 50 µL containing 5 µL 10 × buffer plus MgCl2, 2 mM dNTP, 2 unit Taq DNA polymerase, 100 ng genomic DNA as a template, and 25 picomoles of each primer (luk-PV-1, 5'-ATCATTAGGTAAAATGTCTGGACATGATCCA-3’ and luk-PV-2, 5'-GCATCAASTGTATTGGACATGATCCA-3'). The PCR was performed using a thermal cycler (Eppendorf Mastercycler 5330, Eppendorf-Nethel-Hinz GmbH, Hamburg, Germany) under the following conditions: an initial denaturation for one minute at 94°C and 30 cycles including 94°C for 30 seconds, 45°C for 30 seconds and 72°C for one minute, and a final extension at 72°C for eight minutes.
3.7. Detection of Virulence Genes in The Methicillin Resistant Staphylococcus aureus
The PCR reaction was performed based on a set of primers described previously (23-27). Oligonucleotide primers and PCR programs used for amplification of virulence genes are shown in Table 3. The PCR reactions were performed in a total volume of 30 μL, including 1.8 μL of MgCl2 (25 mmol/ L), 0.6 μL of deoxynucleoside triphosphate (10 mmol/L), 2.5 μL of DNA, 3 μL of 10X PCR buffer, 0.1 μL of Taq DNA polymerase (5 U/μL) and 20 μL of distilled water.
| Types of Clinical Infections | No. Samples Collected | No. MRSA, % | PVL Positive, % |
|---|---|---|---|
| Khorasan | |||
| Blood | 10 | 1 (10) | - |
| UTIs | 10 | 2 (20) | 1 (50) |
| RTIs | 12 | 3 (25) | 2 (66.66) |
| Wound infections | 11 | 3 (27.27) | 1 (33.33) |
| Burn infections | 13 | 4 (30.76) | 2 (50) |
| Total | 56 | 13 (23.21) | 6 (46.15) |
| Tehran | |||
| Blood | 10 | 1 (10) | - |
| UTIs | 14 | 4 (28.57) | 2 (50) |
| RTIs | 15 | 5 (33.33) | 3 (60) |
| Wound infections | 15 | 5 (33.33) | 3 (60) |
| Burn infections | 12 | 4 (33.33) | 3 (75) |
| Total | 66 | 19 (28.78) | 11 (57.89) |
| Isfahan | |||
| Blood | 10 | 1 (10) | - |
| UTIs | 16 | 5 (31.25) | 3 (60) |
| RTIs | 15 | 6 (40) | 4 (66.66) |
| Wound infections | 13 | 5 (38.46) | 3 (60) |
| Burn infections | 13 | 5 (38.46) | 4 (80) |
| Total | 67 | 22 (32.83) | 14 (63.63) |
| Shiraz | |||
| Blood | 10 | 2 (20) | 1 (50) |
| UTIs | 20 | 7 (35) | 4 (57.14) |
| RTIs | 13 | 6 (46.15) | 4 (66.66) |
| Wound infections | 11 | 5 (45.45) | 3 (60) |
| Burn infections | 12 | 5 (41.66) | 4 (80) |
| Total | 66 | 25 (37.87) | 16 (64) |
| Total | |||
| Blood | 40 | 5 (12.5) | 1 (20) |
| UTIs | 60 | 18 (30) | 10 (55.55) |
| RTIs | 55 | 20 (36.36) | 13 (65) |
| Wound infections | 50 | 18 (36) | 10 (55.55) |
| Burn infections | 50 | 18 (36) | 13 (72.22) |
| Total | 255 | 74 (29.01) | 47 (63.51) |
Total Distribution of Methicillin Resistant Staphylococcus aureus in Various Types of Clinical Infections in Pediatric Patients of Four Major Provinces of Irana
3.8. Gel Electrophoresis
Fifteen microliters of PCR products in all reactions were resolved on 2% agarose gel containing 0.5 mg/ml of ethidium bromide in Tris–borate–EDTA buffer at 90 V for one hour, using suitable molecular weight markers. The products were examined under ultraviolet illumination.
3.9. Statistical Analysis
The results were transferred to a Microsoft Excel spreadsheet (Microsoft Corp., Redmond, WA) for analysis. Statistical analysis was performed using the SPSS/16.0 software (SPSS Inc., Chicago, IL) to determine significant relationships between the prevalence of bacteria, PVL genes, SCCmec types and antibiotic resistance pattern of MRSA isolated from various types of clinical infections in pediatric patients. The chi-square test and Fisher’s exact two-tailed test were performed in this study. Statistical significance was regarded at a P value < 0.05.
4. Results
Total distribution of MRSA in various types of clinical infections of pediatric patients from four major provinces of Iran is shown in Table 3. Of the 255 studied samples, 74 (29.01%) were positive for MRSA. Of the 74 MRSA strains, 47 (63.51%) were PVL positive. Our results showed that Shiraz had the highest prevalence of MRSA (37.87%), while Khorasan had the lowest prevalence (23.21%). Respiratory tract infections (36.36%) were the most common infections amongst our samples, followed by burn infections (36%) and wound infections (36%). A significant difference was seen between the types of infections and incidence of MRSA (P = 0.044). Total distribution of antibiotic resistance genes in the MRSA strains of various clinical infections of pediatric patients is shown in Table 4. The most commonly detected antibiotic resistance genes in the MRSA strains of pediatrics clinical infections were tetK (89.18%), mecA (71.62%), msrA (56.75%) and tetM (54.05%). The MRSA strains of burn infections had the highest prevalence of antibiotic resistance genes. A significant difference was seen between the types of infections and incidence of antibiotic resistance genes (P = 0.038).
| Types of Infections (No. MRSA) | Distribution of Antibiotic Resistance Genes, % | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mecA | aacA-D | tetK | tetM | msrA | msrB | vatA | vatB | vatC | ermA | ermC | linA | |
| Blood (5) | 1 (20) | 1 (20) | 4 (80) | 2 (40) | 4 (80) | 2 (40) | 1 (20) | - | - | 2 (40) | 1 (20) | 1 (20) |
| UTIs (18) | 8 (44.44) | 3 (16.66) | 13 (72.22) | 7 (38.88) | 6 (33.33) | 2 (11.11) | 1 (5.55) | 1 (5.55) | - | 3 (16.66) | 1 (5.55) | 2 (11.11) |
| RTIs (20) | 15 (75) | 4 (20) | 16 (80) | 10 (50) | 9 (45) | 4 (20) | 4 (20) | 2 (10) | 1 (5) | 5 (25) | 2 (10) | 4 (20) |
| Wound infections (18) | 14 (77.77) | 5 (27.77) | 16 (88.88) | 10 (55.55) | 11 (61.11) | 6 (33.33) | 4 (22.22) | 1 (5.55) | 1 (5.55) | 4 (22.22) | 2 (11.11) | 4 (22.22) |
| Burn infections (18) | 15 (83.33) | 5 (27.77) | 17 (94.44) | 11 (61.11) | 12 (66.66) | 6 (33.33) | 2 (11.11) | - | - | 6 (33.33) | 3 (16.66) | 5 (27.77) |
| Total (74) | 53 (71.62) | 18 (24.32) | 66 (89.18) | 40 (54.05) | 42 (56.75) | 20 (27.02) | 12 (16.21) | 4 (5.40) | 2 (2.70) | 20 (27.02) | 9 (12.16) | 16 (21.62) |
Total Distribution of Antibiotic Resistance Genes in the Methicillin Resistant Staphylococcus aureus Strains of Various Clinical Infections of Pediatric Patients
Susceptibility of MRSA strains against 18 commonly used antimicrobial agents is shown in Table 5. The MRSA strains of various clinical infections of pediatric patients harbored the highest levels of resistance against penicillin (100%), tetracycline (98.64%), ampicillin (93.24%) and oxacillin (93.24%). The MRSA strains of burn and wound infections had the highest prevalence of resistance against the tested antimicrobial agents. A significant difference was also seen between the types of infections and incidence of antibiotic resistance (P = 0.026).
| Antimicrobial Agents | Types of Infections, % | |||||
|---|---|---|---|---|---|---|
| Blood (5) | UTIs (18) | RTIs (20) | Wound infections (18) | Burn infections (18) | Total (74) | |
| Ampicillin | 4 (80) | 15 (83.33) | 17 (85) | 16 (88.88) | 17 (94.44) | 69 (93.24) |
| Gentamycin | - | 2 (11.11) | 6 (30) | 8 (44.44) | 8 (44.44) | 24 (32.43) |
| Lincomycin | - | - | 1 (5) | 2 (11.11) | 2 (11.11) | 5 (6.75) |
| Cephalothin | - | 3 (16.66) | 2 (10) | 1 (5.55) | 2 (11.11) | 8 (10.81) |
| Imipenem | - | 1 (5.55) | - | 1 (5.55) | - | 2 (2.70) |
| Tetracycline | 4 (80) | 16 (88.88) | 18 (90) | 17 (94.44) | 18 (100) | 73 (98.64) |
| Vancomycin | 1 (20) | 1 (5.55) | 1 (5) | 1 (5.55) | 1 (5.55) | 5 (6.75) |
| Ciprofloxacin | 1 (20) | 4 (22.22) | 3 (15) | 5 (27.77) | 5 (27.77) | 18 (24.32) |
| Norfloxacin | 1 (20) | 5 (27.77) | 5 (25) | 6 (33.33) | 7 (38.88) | 24 (32.43) |
| Cotrimoxazole | 1 (20) | 2 (11.11) | 2 (10) | 3 (16.66) | 4 (22.22) | 12 (16.21) |
| Clindamycin | 1 (20) | 2 (11.11) | 3 (15) | 3 (16.66) | 3 (16.66) | 12 (16.21) |
| Trimethoprim-sulfamethoxazole | 1 (20) | 3 (16.66) | 6 (30) | 7 (38.88) | 9 (50) | 26 (35.13) |
| Penicillin | 4 (80) | 17 (94.44) | 18 (90) | 17 (94.44) | 18 (100) | 74 (100) |
| Oxacillin | 3 (60) | 15 (83.33) | 17 (85) | 16 (88.88) | 18 (100) | 69 (93.24) |
| Erythromycin | 2 (40) | 10 (55.55) | 12 (60) | 13 (72.22) | 15 (83.33) | 42 (56.75) |
| Azithromycin | 2 (40) | 8 (44.44) | 9 (45) | 12 (66.66) | 13 (72.22) | 44 (59.45) |
| Ceftriaxone | 1 (20) | 2 (11.11) | 3 (15) | 5 (27.77) | 5 (27.77) | 16 (21.62) |
| Cefixime | 1 (20) | 2 (11.11) | 5 (25) | 7 (38.88) | 7 (38.88) | 22 (29.72) |
Susceptibility of Methicillin Resistant Staphylococcus aureus Strains of Various Types of Clinical Infections in Pediatric Patients Against Commonly Used Antibiotics
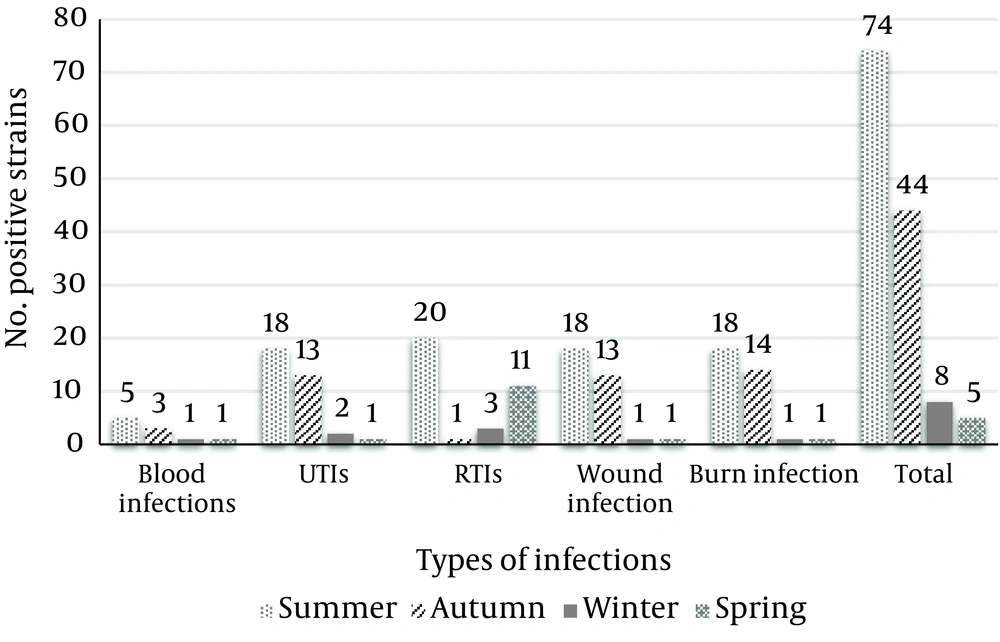
Total distribution of SCCmec types in the MRSA strains of various clinical infections of pediatric patients is shown in Table 6. Our results showed that the most commonly detected SCCmec types in the MRSA strains were type V (18.91%) and III (17.56%). Figure 1 shows the seasonal distribution of MRSA in various types of clinical infections of pediatric patients. The MRSA strains of blood, UTIs, wound and burn infection sources had the highest prevalence in the summer season, while those of RTIs had the highest prevalence in the winter season.
| Types of Infections (No. MRSA) | Distribution of SCCmec Types, % | |||||||
|---|---|---|---|---|---|---|---|---|
| I | II | III | IVa | IVb | IVc | IVd | V | |
| Blood (5) | - | - | - | 1 (20) | - | - | - | 1 (20) |
| UTIs (18) | 1 (5.55) | - | - | 1 (5.55) | - | - | - | 2 (11.11) |
| RTIs (20) | 2 (10) | 1 (5) | 2 (10) | 2 (10) | - | 2 (10) | 1 (5) | 3 (15) |
| Wound infections (18) | 1 (5.55) | 1 (5.55) | 5 (27.77) | 4 (22.22) | 1 (5.55) | 1 (5.55) | 3 (16.66) | 4 (22.22) |
| Burn infections (18) | 1 (5.55) | 1 (5.55) | 6 (33.33) | 4 (22.22) | 1 (5.55) | 2 (11.11) | 3 (16.66) | 4 (22.22) |
| Total (74) | 5 (6.75) | 3 (4.05) | 13 (17.56) | 12 (16.21) | 2 (2.70) | 5 (6.75) | 7 (9.45) | 14 (18.91) |
Total Distribution of Staphylococcal Chromosomal Cassette mec Types in the Methicillin Resistant Staphylococcus aureus Strains of Various Clinical Infections of Pediatric Patients
5. Discussion
The present investigation focused on the study of the prevalence of MRSA in various types of clinical infection samples of pediatric patients as well as the distribution of PVL gene, SCCmec types and antibiotic resistance genes of the MRSA isolates. Study of the seasonal and geographical distribution of MRSA was the other purpose of the present investigation. Our results showed that 29.01% of the clinical infection samples were positive for MRSA. This maybe due to the fact that the prescription of methicillin is very high in Iranian health centers and hospitals. In addition, this finding showed that the Iranian hospital’s environments are highly infected. Also, our results showed that antibiotics were used in a highly irregular manner in Iranian hospitals. Therefore, it is not surprising that the prevalence of resistance against penicillin, tetracycline, ampicillin and oxacillin were 100%, 98.64%, 93.24% and 93.24%, respectively. These findings may lead to the emergence of resistant staphylococcal diseases, which can infect patients and even healthy people in hospital environments. Similar results have been reported previously (2, 3, 5).
A study on Lebanese hospitals by Tokajian et al. (6) reported that the prevalence of MRSA was 72%. They showed that 18% of the MRSA strains were resistant to 10 - 18 antibiotics. Udo et al. (8) revealed that 32% of inpatients and outpatients of Kuwait hospitals were positive for MRSA. High prevalence of MRSA in various types of clinical infections was also reported by Alghaithy et al. (61% in Saudi-Arabia) (28), Młynarczyk et al. (40% in Warszawie) (29) and Rijal et al. (56.1% in Pokhara) (30). Virdis et al. (7) showed that the prevalence of resistance of S. aureus against kanamycin, oxytetracycline and ampicillin was 28%, 16% and 12%, respectively. Deng et al. (31) reported on the high prevalence of resistance of S. aureus against most commonly used antibiotics including nafcillin, oxacillin, vancomycin and cefathiamidine. Nishijima and Kurokawa (32) showed that the prevalence of S. aureus resistance against penicillin, cephalosporins and clindamycin was 20 to 30%. They showed that the prevalence of resistance against gentamycin, erythromycin and methicillin was 55.2%, 39.6% and 21%, respectively. Kumar et al. (33) revealed that S. aureus isolates of clinical infections were highly resistant to different antibiotics, i.e. 33.6% were resistant to oxytetracycline, 36.4% to streptomycin, 29.9% to gentamycin and 26.2% to chloramphenicol, pristinamycin and ciprofloxacin; these findings are similar to our results.
The most effective antibiotics against MRSA strains of our investigation were imipenem, lincomycin, vancomycin, cephalothin, cotrimoxazole and clindamycin yet similar results showing a higher efficacy for imipenem against MRSA strains of human clinical infections have been reported by other investigations (34). Boyce et al. (35) reported that the prevalence of resistance against imipenem in the MRSA strains of clinical infections was 57%, which was higher than our results. There was low bacterial resistance against vancomycin in our study yet high resistance against vancomycin has been reported previously (36-38). Current recommendations indicate that antibiotics may trigger release of PVL and progression to negative clinical complications. The results of the disk diffusion method were also confirmed by the PCR technique. The most commonly detected antibiotic resistance genes in the MRSA strains were tetK (89.18%), mecA (71.62%), msrA (56.75%) and tetM (54.05%). Momtaz and Hafezi (39) showed that S. aureus strains of hospital infections harbored mecA (80.30%), tetK (34.84%), ermA (30.30%), vatB (1.51%) and vatC (3.03%) antibiotic resistance genes, which was similar to our results. A Nigerian study reported that mecA, aacA-ophD and tetM were the most common antibiotic resistance genes in cases of UTIs (40). They showed that the ermA gene was identified in all erythromycin-resistant MRSA isolates, while two erythromycin-resistant methicillin sensitive staphylococcus aureus (MSSA) isolates possessed the msrA gene. Besides, all the gentamicin-resistant isolates carried the aacA-aphD gene. Moreover, the tetM gene was detected in 11 isolates (seven MRSA and four MSSA) and the tetK gene was present in four MRSA and 23 MSSA isolates (40).
Overall, 63.51% of the MRSA strains of our investigation were PVL positive. This gene had the highest incidence in burn infections (72.22%), RTIs (65%) and wound infections (55.55%) of our study. The main reason for the high incidence of PVL gene in wound and burn infections and RTIs is the fact that this gene is a bicomponent cytotoxin that is linked to furuncles, cutaneous abscess and severe necrotic skin and respiratory infections. Significant statistical differences were seen in the distribution of the PVL gene between various types of infections (P < 0.05). The PVL gene was found in 80% of cutaneous abscesses, 33.33% of finger pulp infections, 100% of cases with pneumonia and 100% of cases with furunculosis (41). High prevalence of the PVL gene in the S. aureus strains of human clinical infections have been reported by Khosravi et al. (Iran) (7.23%) (42), Esan et al. (Nigeria) (18%) (43) and Shallcross et al. (United Kingdom) (11.3%) (44). The total prevalence of the PVL gene has been estimated to be 2 to 35% among MRSA strains in various types of hospital infections (45, 46). The PVL toxin’s ability to cause death of polymorphonuclear cells including neutrophils, basophils and eosinophils has been known since its earliest descriptions.
One of the most interesting findings of our investigation was determination of differences in the prevalence of MRSA in various types of infections in different seasons. Summer had the highest, while spring had the lowest prevalence of MRSA. Cold and windy climate in the winter season is the main reason for the higher prevalence of MRSA in cases of RTIs in the winter season. Coldness and windy climate can easily damage the respiratory tract. The main reason for the higher prevalence of MRSA in summer season in Iran is the fact that during this period, climatic events, heat, rain, and thunderstorms, as well as variation of barometric pressure may have influences on the autonomic nervous system. These events cause reduction in the levels of human immunity. Therefore, several infections might occur. In keeping with this, the MRSA bacteria have higher growth and surveillance in warm conditions. The levels of public and individual health are also decreased in warm climates like summer. After analyzing the average temperatures of these four seasons in the study area (16°C for spring, 36°C for summer, 13°C for autumn and 4°C for winter), it was recognized that the prevalence rate of MRSA strains in each season is related to average temperatures. A significant difference (P = 0.029) was seen between the average temperatures of hot and cold seasons. In the field of geographical distribution, Shiraz due to its high temperature and moisture had the highest prevalence of MRSA, followed by Isfahan.
The V (18.91%) and III phenotypes (17.56%) were the most commonly detected SCCmec types among the MRSA strains of our study. Similar studies have reported a higher prevalence of SCCmec V and III types in various clinical infections (21, 47-49). High prevalence of SCCmec IV type in various types of infections has also been reported by D’Souza et al. (48) and Moussa et al. (2). In conclusion, we identified a large number of MRSA, PVL gene and antibiotic resistance properties in the MRSA strains of hospitalized infection samples of pediatric patients. Our data indicated that MRSA strains are predominant in Iran. High PVL gene distribution was also found. Our data revealed that tetK, mecA, msrA and tetM resistance genes and resistance against penicillin, tetracycline, ampicillin and oxacillin were the most commonly detected characteristics of the MRSA strains isolated from hospitalized infection samples of pediatric patients. High prevalence of MRSA, high levels of resistance against commonly used antibiotics as well as presence of SCCmec types indicate that infections with these strains require higher levels of hospital care with high demand for novel antibiotics. Hence, judicious use of antibiotics is required by clinicians. Shiraz due to its high temperature and moisture had the highest prevalence of MRSA. Simultaneous presence of PVL and SCCmec genes with high prevalence of resistance against commonly used antibiotics in some strains of MRSA strains in infected children indicated an important public health problem. Prescription of imipenem, lincomycin and cephalothin can be effective for treatment of cases of infection due to MRSA strains.