1. Background
Malassezia species are lipophilic or lipid-dependent, budding yeasts, which are part of the skin microflora of humans and other warm-blooded animals. It can become pathogenic under certain predisposing factors, such as alterations in the skin’s condition and changes in the host’s defenses. Malassezia species are associated with various human diseases, especially pityriasis versicolor (PV), which is a chronic superficial skin disorder (1). Pityriasis versicolor is a superficial fungal infection characterized by the appearance of round to oval lesions. PV is a chronic cutaneous disorder, and it can occur in any part of the word; however, it is more common in tropical, humid areas. Despite antifungal treatment, most patients with PV will face recurrence of the condition.
The disease is more common between puberty and middle age, when there are maximum amount of Malassezia yeast on the skin. PV is characterized by a low to more scaly macule that can present as either hyper or hypopigmented (1), and it is usually present on the neck and upper trunk (2). Moreover, Malassezia species are an etiological agent of catheter-associated; fungemia, seborrheic dermatitis (3), folliculitis (1, 4), dacryolitis, and blepharitis, as well as nosocomial bloodstream infections in pediatric care units (5).
In recent years, molecular studies have changed the taxonomy of the genus Malassezia to a considerable extent. Sequence variability between Malassezia species has been documented in rRNA genes. In 1996, Gueho et al. (6), classified the genus of Malassezia in seven distinct species, namely; M. furfur, M. pachydermatis, M. sympodialis, M. globosa, M. obtusa, M. restricta and M. slooffiae. Recently, based on DNA analysis, six new species have been introduced: M. dermatis, M. nana, M. japonica, and M. yamatoensis, M. equine, M. caprae. Of these M. caprae, M. equine and M. nana have only been isolated from domestic animals (6-10).
Malassezia species can be diagnosed based on their biochemical features. However, these methods do not have enough discriminatory power and therefore, cannot characterize the newly defined species. In addition, biochemical and phenotypical methods are not able to achieve an immediate diagnosis. In recent years, molecular approaches and PCR methods for the discrimination of Malassezia species are the most accurate (11, 12). Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) using restriction enzyme digestion is a simple, reliable, and cost-effective method for the differentiation of Malassezia species, which has been described by Mirhendi et al. (13). After amplification of the partial 26S rDNA, digestion of the PCR product with the enzyme CfoI produced the predicted species specific pattern for all species tested. Using CfoI, nine different species, including; M. furfur, M. pachydermatis, M. globosa, M. obtuse, M. restricta, M. slooffiae, M. nana, M. japonica, and M. yamatoensis could be distinguished, however, M. sympodialis and M. dermatis produced the same digestion pattern.
Our data offers insight into the epidemiology of PV in Iran, but it is limited to areas such as; Tehran, Ahvaz, Yazd and Mazandaran (4, 14-17). Furthermore, there is evidence suggesting geographical variations in the distribution of the species. On the other hand, the majority of these studies have identified Malassezia species based on biochemical and phenotypical characterization, such as assimilation of Tween, and PCR based techniques were not used.
2. Objectives
The objective of this study was to determine the Malassezia species which grew on culture from the skin scrapings of patients with PV in Kashan (central region of Iran), using molecular methods. There is little current information about the molecular epidemiology and ecology of Malassezia species available in Kashan, Iran.
3. Patients and Methods
3.1. Subjects and Collecting Samples
In total, 140 subjects with skin lesions suspected to be PV from the Shahid Beheshti Skin Clinic, during 2012, who were referred to the Fatemieh Zeidi Clinical Laboratory, Kashan, Iran, were enrolled in this study. A data collection form which included; sex, age, disease duration, and lesion area, was completed for the patients. Identification of PV on the lesions was initially clinically diagnosed after that the final diagnosis was confirmed by direct microscopic examination using the Sellotape method and methylene blue stain for skin samples, obtained by the scraping method (18, 19).
3.2. Culture of Sample
The scraped skins were inoculated in modified Dixon medium. This medium consisted of 3.6% malt extract (Merck, Germany), 2.0% desiccated ox-bile, 1.0% Tween 40 (Sigma & Aldrich), 0.2% glycerol (Merck, Germany), 0.2% oleic acid (Merck, Germany), 0.05% chloramphenicol (Sigma, USA), 0.5% cycloheximide (Sigma, USA), and 1.2% agar. Inoculated cultures were incubated at 32ºC for 1-10 days. The harvested cells were preserved at -20˚C until diagnostic analysis. Suspected Malassezia species were identified to the level of species on the basis of the PCR-RFLP method (13).
3.3. PCR- RFLP
DNA was extracted with phenol chloroform-isoamyl alcohol as described by Gupta et al. (20). The PCR reaction was performed in a final volume of 50 µL. Each reaction contained 1 µL of DNA template, 0.5 µL of each primer, 0.20 mm of each deoxynucleoside triphosphate (dNTPs), 5 µL of 10X PCR buffer, and 1.25 U of Taq polymerase, using 2720 Applied Biosystem thermocycler (life technologies, US). The primer nucleotides was as follows: forward, 5´TAACAAGGATTCCCCTAGTA-3´, and reverse 5´-ATTACGCCAGCATCCTAAG-3´. After initial denaturation at 94˚C for 5 minutes, the reaction was followed by 30 cycles of denaturation at 94˚C for 45 seconds, annealing at 55˚C for 1 minute, and extension at 72˚C for 45 seconds, with a final extension step of 72˚C for 7 minutes.
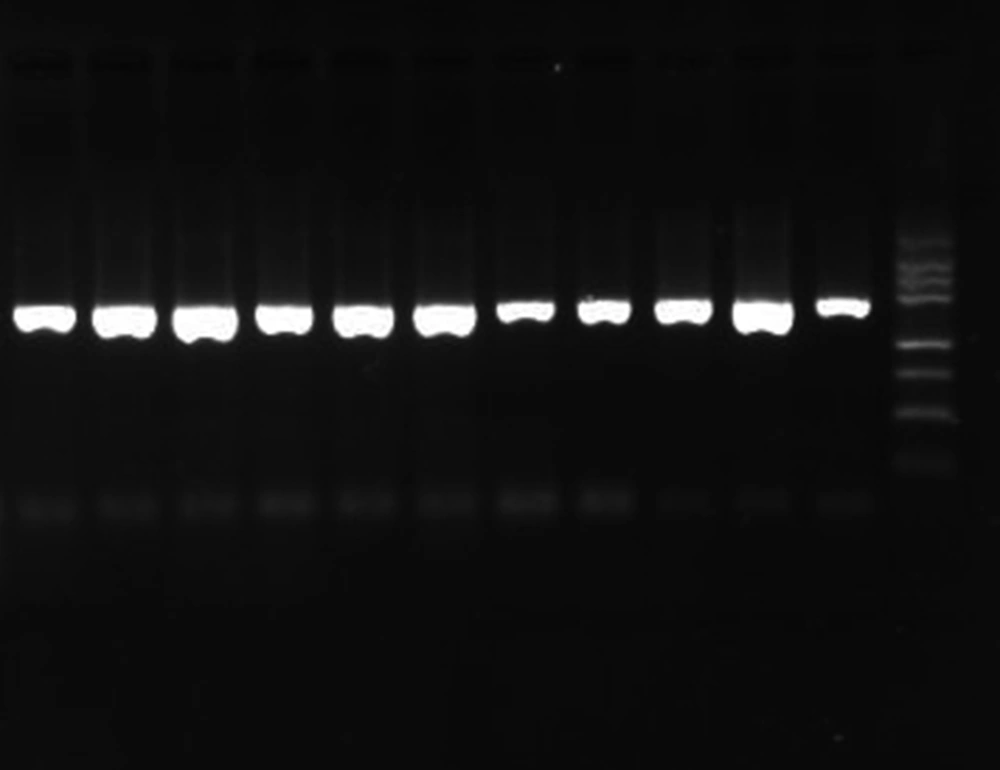
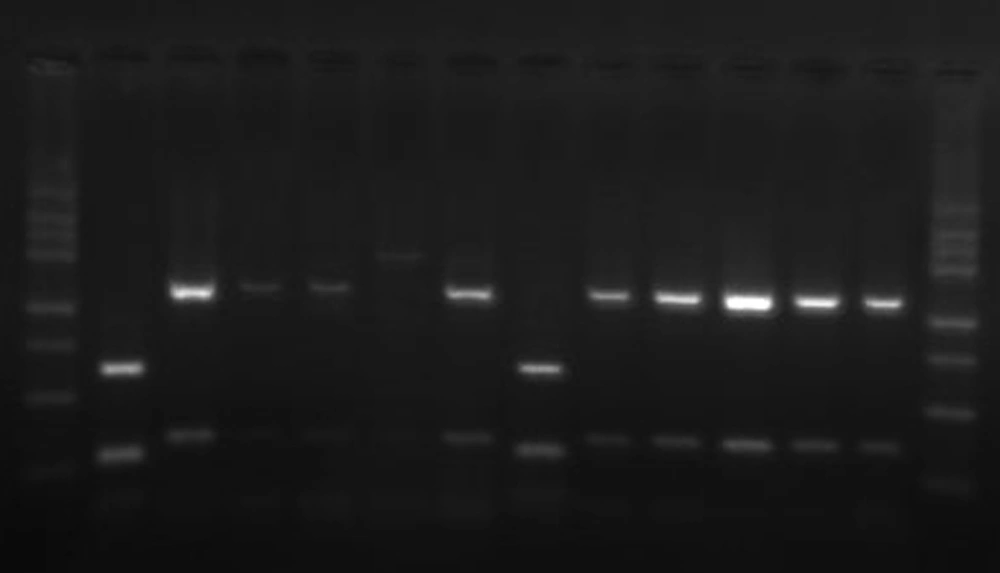
PCR products were evaluated by 1.5% (w/v) agarose gel electrophoresis in tris-boric acid-EDTA (TBE) buffer, stained with ethidium bromide, and photographed under UV transillumination. The target part of the 26rDNA region was amplified for the Malassezia isolates, to produce a PCR product approximately 580bp in length (Figure 1). PCR products were used for further restriction fragment length polymorphism (RFLP) by CfoI (Roche, Germany) enzymes. Digestion was performed by incubating a 20 µL aliquot of PCR products with 10 U of the enzyme in a final reaction volume of 25 µL at 37°C for 3 hours, followed by 2% ethidium bromide agarose gel electrophoresis in a TBE buffer (Figure 2).
4. Results
Direct examination of the specimens was positive in 93.3% (112/120) of questionable PV lesions. However, only 89.3% (100/112) of the positive specimens yielded Malassezia in modified Dixon agar. The patients with PV included 60 men and 52 women, aged between 1 and 85 years. Statistically there was no significant difference regarding gender and nor was it observed in the frequency of Malassezia species isolated between women and men in each group or between groups (P = 0.3). The average age of the patients was 26 years. The prevalence of PV in patients 21–30 years-of-age was higher than in the other age groups (Table 1). In the present study, no difference was seen in the frequency of Malassezia species occurrence depending on the age of the patients. In total, 65% of patients with PV had hyperhidrosis. In 86% of patient samples, yeast and hyphal forms of Malassezia were observed in direct microscopy examination of PV lesions.
M. globosa (66%) was the most frequent Malassezia species isolated in the PV lesions followed by M. furfur (26%), M. restricta (3%), M. sympodialis (3%), and M. slooffiae (2%), respectively. Overall, M. globosa and M. furfur were the most frequently isolated organisms. The prevalence of Malassezia species based on the gender of patients is shown in Table 2. Malassezia species were mainly isolated from the neck and chest. Overall, 36 and 26 isolates were obtained from these sites, respectively. In addition, the lowest number of Malassezia species isolated were from the arms and abdomen (Table 3).
A statistical analysis of isolated Malassezia species showed that there was no significant relationship between Malassezia species and any of the anatomical sites (P values = 0.2). The distribution of sites and Malassezia species based on pigmentation, is shown in Table 4. The duration of the disease ranged less than six months in 44% of patients, followed by; six months to one year (26%), one to two years (10%), and over two years (20%).
| Age Group | Malassezia Species (n) | |||||
|---|---|---|---|---|---|---|
| M. globosa | M. furfur | M. restricta | M. sympodialis | M. slooffiae | Total | |
| 1-10 y | 6 | 5 | 0 | 0 | 0 | 11 |
| 11-20 y | 15 | 5 | 1 | 1 | 0 | 22 |
| 21-30 y | 25 | 6 | 1 | 1 | 2 | 35 |
| 31-40 y | 11 | 5 | 1 | 0 | 0 | 17 |
| 41-50 y | 5 | 3 | 0 | 1 | 0 | 9 |
| > 50 y | 4 | 2 | 0 | 0 | 0 | 6 |
| Total | 66 | 26 | 3 | 3 | 2 | 100 |
Frequency of Malassezia Species According to Age Groups a
| Skin Pigmentation | Malassezia Species | |||||
|---|---|---|---|---|---|---|
| M. globosa | M. furfur | M. restricta | M. sympodialis | M. slooffiae | Total | |
| Hyperpigmentation | 31 | 15 | 2 | 1 | 1 | 50 |
| Hypopigmentation | 25 | 10 | 1 | 0 | 1 | 37 |
| Both | 10 | 1 | 0 | 2 | 0 | 13 |
| Total | 66 | 26 | 3 | 3 | 2 | 100 |
Distribution of Malassezia Species Isolated According to Clinical Signs a
| Species | Anatomical Sites (n) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Neck | Back | Chest | Face | Arm | Trunk | Limb | Groin | Total | |
| M. globosa | 25 | 20 | 17 | 14 | 12 | 10 | 5 | 4 | 107 |
| M. furfur | 8 | 5 | 6 | 6 | 3 | 4 | 2 | - | 34 |
| M. restricta | 2 | 1 | 1 | - | - | - | - | - | 4 |
| M.sympodialis | 1 | 1 | 1 | - | - | - | - | - | 3 |
| M.slooffiae | - | - | 1 | - | - | 1 | - | - | 2 |
| Total | 36 | 27 | 26 | 20 | 15 | 15 | 7 | 4 | 150 |
Distribution of Malassezia species Isolated Based on Lesion Area
Lanes 2, 8: M. furfur, with 250, two ~107-113bp (as overlapping) fragments, lanes 3-5, 7, 9-13: M. globosa, with 129 and 455bp fragments, lane 6: M. restricta, with 581bp fragments, lane 1, 14:100 bp ladder. M. furfur has multiple fragments (59, 30, 21, 2bp) not distinguishable after gel electrophoresis.
5. Discussion
The genus of Malassezia consists of at least thirteen different species, which have been identified based on biochemical and morphological features. However, biochemical and morphological features often do not allow the rapidly and exact characterization of related Malassezia species, so molecular approaches need to be used in studies of the epidemiology of Malassezia, and pathogenesis and disease caused by the Malassezia genus.
The present study used the PCR-RFLP method for the identification of the Malassezia genus in order to determine the specific identification of Malassezia species and the diagnosis of related infections. M. globosa and M. furfur were isolated significantly more often than other species in this study; albeit at unequal rates, as M. globosa was isolated from 66% of patients with PV. Recent studies from Iran have showed that M. globosa is the commonest Malassezia species isolated from Malassezia skin disorders. In contrast to our results, others studies from Iran have reported lower rates of isolation of M. globosa as predominant in different geographical areas at the rate of 41-47.6 (4, 14, 16, 19). In addition, compatible with present studies, M. globosa are the major causative agent of PV in other countries (21, 22).
Hedayati et al. (18) reported 58% M. globosa from patients with Seborrheic dermatitis. Zomorodian et al. (15), reported that M. furfur is the most common Malassezia species isolated from psoriasis and healthy individuals. Some investigators from other countries have reported M. globosa as the prevailing Malassezia species correlated to PV, while others report M. sympodialis or M. furfur as the species with the highest rates of isolation from PV lesions, however, we found no M. sympodialis in our findings. In accordance with our data M. slooffiae and M. restricta were less commonly isolated species as found in previous studies (23).
On the other hand, similar to other published data, the highest prevalence of PV in our study was observed in the 21–30 year-old group, this is likely to be the result of increased sebum discharge at these ages (4, 24). Previous studies have shown that PV is less common in children (4, 14, 25), but we found 11 cases of PV in children, and this may be due to climate and geographical conditions. Kashan is located on the border of a desert, and as a result it has a hot climate that facilitates the occurrence of Malassezia skin infections. Giusiano et al. (25) also reported 9% and 24% of children with Malassezia infections in the under 5 and up to 15 years, respectively. Different studies have concluded that PV is rarely found in the aged individual, and we had lower cases of PV in the over 50 year old group.
We found no isolates of M. obtusa and M. pachydermatis in this study, although in other studies in Iran these species were isolated at a low frequency. In previous reports from Iran, M. pachydermatis was not isolated, or it was isolated at a low frequency (8.5%). These differences in the prevalence of Malassezia species between our study and other reports may be due to geographical variation and some laboratory techniques such as sampling and diagnostic methods. Racial factors and geographical location may be factors in these differences (11). We have isolated single separated colonies from each lesion site, as suggested by some investigators in patients with PV. However, different studies have isolated more than one species from each sample. Obtaining pure culture from mixed samples is usually very difficult, Due to the fact that, some fast and simple growing Malassezia species inhibit other species in the culture.
PV lesions were frequently observed on the trunk and arms (26, 27). The high frequency of PV lesions on the neck in our study is controversial, we also observed a number of patients with multiple lesions (30%). On the other hand, some patients with lesions in unexpected locations, such as the face and limbs, and especially the groin, were diagnosed. The higher ambient temperature of our region probably encourages the spread of PV. Epidemiological studies have mentioned the distribution of some species in defined anatomical sites (27). We observed no differences in the frequency of Malassezia species based on the anatomical sites, which might be related to the high number of PV patients. Our findings were compatible to those of other similar studies (17, 25). In this study, the recovery rate of Malassezia species from PV lesions was 89.3%. In similar studies, this ratio was different and ranged between 45-90% (4, 17, 18, 22, 27).
In the new taxonomy by Gueho et al. (6), different studies have evolved with the purpose of clarifying the epidemiology and role of the Malassezia species in skin disorders. This work will help improve knowledge of Malassezia genus epidemiology, especially in the central areas of Iran. Future studies will be necessary in order to explain the ecology and relationships of these species with human disease. Molecular techniques have been useful in the identification and discrimination of Malassezia species and this provides information about their epidemiology. By using these methods, the detection and identification of individual Malassezia species from clinical samples is made substantially easier.