1. Background
Antibiotics are usually assumed as secondary metabolites produced during the idiophase of microbial growth. Nowadays, the rapid spread of multidrug-resistant bacteria, re-emergent infections, and newly emerging infectious diseases have become a serious global public health threat, specifically affecting the economic development (1, 2). Multi antibiotic-resistant bacteria are serious health challenges in hospital environments. They become recalcitrant to antimicrobial therapy. Therefore, there is a need for novel antimicrobial agents which are effective against multidrug-resistant bacteria (3). Studying different environments throughout the world has yielded a lot of antimicrobial agents that are of great value for treatment of many infectious diseases. Animals are considerable for these researches, since they defend themselves against pathogenic microorganisms via production and secretion of antimicrobial components.
Among the animals, amphibians are of great interest. The moist surface of frog skin creates a suitable setting for pathogenic microorganisms (4, 5). The symbiotic microorganisms that live on frog skin, influence the health of amphibians and may affect the innate immune defense of the host. Innate skin defense consists of antimicrobial peptides and symbiotic microbials, which may be crucial for protecting the amphibians against skin-invasive pathogens (6).
Reports about isolation of antimicrobial-producing bacteria from frog skin are rare. Woodhams et al. suggested that the symbiotic bacteria on the skin of Rana muscosa that were able to persist in the presence of mucosal peptides, may inhibit the infection and colonization of Batrachochytrium dendrobatidis, a pathogenic chytrid fungus, on the skin, and increase the efficiency of skin innate defense mechanisms (6). Bacteria have been competing for niches for millions of years and have developed elaborate systems to inhibit competitors. The enormous genetic potentials of microorganisms can be used for generating useful compounds. As mentioned, antibiotic resistance of pathogenic bacteria is increasing day by day due to extensive nonmedical usages of antibiotics and drugs. This concern is becoming a challenge for researchers to investigate new antimicrobial agents produced by bacterial strains with low virulence capabilities and positive antibacterial activities against wide range of clinically significant organisms.
2. Objectives
In this study, we investigated the antimicrobial agents produced by a bacterium isolated from the skin of R. ridibunda, an endemic frog in Iran. Identification and characterization of this bacterium as well as extraction of antimicrobial substance were studied.
3. Materials and Methods
3.1. Materials
Forward: 5'-AGAGTTTGATCCTGGCTCAG-3' and Reverse: 5'-TAAGGAGGTGATCCAGCC-3' primers were purchased from Genfanavaran, Iran; 1-kb polymerase chain reaction (PCR) marker from Fermentas, Lithuania; high pure PCR kit from Roche, Germany; nutrient agar and methyl red & Voges-Proskauer (MRVP) media from Oxid, Canada Tris base from Sigma, USA; sulfide, indole, motility (SIM) medium from Difco, England; and glucose, starch, arabinose, calcium chloride, and sodium acetate from Fluka, Switzerland. Other materials used in this study were obtained from the Merck Company.
3.2. Bacterium Strain
The bacterium used in this study was previously isolated from the skin of R. ridibunda in our laboratory according to the standard procedures and was designed as GA strain.
3.3. Identification of Bacterium
Identification and characterization of the GA strain was performed on the basis of the cell and colony morphology, growth characteristics, motility, various staining reactions and various biochemical tests, as given by Bergey’s manual of systematic bacteriology (7). Results of the biochemical tests were further confirmed by molecular tests.
For molecular characterization, the 16SrDNA gene fragment was amplified by PCR. The genomic DNA was extracted through culturing the bacterium overnight, using the high pure PCR purification kit (Roche, Germany). The following thermal cycle profile was used for PCR: 94°C for five minutes, 30 cycles of 94°C for one minute, annealing at 56°C for 40 seconds, 72°C for one minute, and a final extension at 72°C for five minutes. Universal primers FD1: 5'-AGAGTTTGATCCTGGCTCAG-3' and RD1: 5'-TAAGGAGGTGATCCAGCC-3' were used in this process. The PCR product was separated on 1% agarose gel and recovered by the high pure PCR purification kit (Roche, Germany). Nucleotide sequencing analysis was performed with dideoxy chain termination method (SEQLAB, Germany). The sequences were compared with similar sequences of the reference organisms by BLAST algorithm (www.ncbi.nlm.nich.-gov/BLAST/).
3.4. Antimicrobial Activity of the GA Strain
Antimicrobial activity of the culture supernatant was examined against the indicator bacteria by disc diffusion and minimum inhibitory concentration (MIC) methods.
3.4.1. Disk Diffusion Method
The GA strain was grown in a synthetic medium at 35°C and 140 rpm for 72-144 hours and in nutrient broth for 24-48 hours. The synthetic medium contained the following ingredients per liter: glucose, 10; K2HPO4, 0.5; NH4Cl, 1; MgSO4.7H2O, 0.2; FeSO4.7H2O, 0.01; CaCl2.2H2O, 0.01; it also contained 1 mL of trace elements(ZnCl2,70 mg; MnCl2.4H2O, 100 mg; CoCl2.6H2O, 200 mg; NiCl2.6H2O, 100 mg; CuCl2.2H2O, 20 mg; NaMoO4.2H2O, 50 mg; Na2SeO3.5H2O, 26 mg; NaVO3.H2O, 10 mg; Na2WO4.2H2O, 30mg; in 1000 mL distilled water)(8). The culture medium was centrifuged at 3400 × g for 20 minutes and pH of the supernatant was adjustment to 7.0. Afterwards, 2 mL of the supernatant was dried on sterile antibiotic assay disks and placed on Muller Hinton agar plates which were previously spread with indicator strains; then, the plates were incubated at 37°C for 18-24 hours and the clear zones were investigated. All the assays were conducted in triplicates.
Standard tested microorganisms were Bacillus subtilis ATCC 465, Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 85327, Candida albicans ATCC 10231, and Aspergillus niger (isolated in our lab). B. subtilis ATCC 465 was used as the indicator strain in subsequent experiments because the antimicrobial compound produced the maximum mean diameter of inhibition zone against this bacterium.
3.4.2. Micro Dilution Assays
The MIC was also studied for antimicrobial assay. The culture supernatant was first washed with chloroform and the raffinate was extracted with methanol. The methanol extract was dried by a vacuum pump at 30°C. The resulted powder was dissolved in distilled water. Then, except for the first well, 250 μL of sterile Muller Hinton broth was poured in each well of a 96-well microtiter plate (Greiner, Nurtingen, Germany). Five milligrams of dried powder was dissolved in the first well and mixed well. Then, 100 µL of this well content was added to the second well and this manner was continued to the penultimate well; the last well was used as the negative control. The inocula of the microorganisms were prepared from the 12-hour broth cultures and finally 100 μL of the standard bacteria suspension, adjusted to the 0.5 McFarland standard turbidity, was added to each well. The plate was incubated for 18-24 hours at 37°C. The lowest concentration that completely inhibited the growth of the bacterial cells was defined as the MIC.
3.5. Effects of Different Media on Antimicrobial Production
To select a culture medium that could enable the optimal production of antimicrobial activities, several liquid culture media, including nutrient broth, Muller Hinton broth and synthetic media, with various carbon sources, including glucose, glycerol and sodium acetate, were tested. The isolated bacterium was inoculated at a 250-mL flask containing 100 mL of each medium and incubated in shaker incubator at 37°C and pH = 7.0. Daily, the cell growth was checked and pH regulating was performed. About 2 mL of the aliquots was collected regularly to estimate the antibiotic activity against B. subtilis by agar disk diffusion method.
3.6. Effects of Temperature and pH on Stability of the Antimicrobial Compound
The antimicrobial activity was tested for sensitivity to heat and pH. To determine the effect of temperature on the antimicrobial compound activity, aliquots of the culture supernatant were prepared in screw capped tubes, adjusted to pH = 7 and placed at 4, 35, 50, 60, 70, 80 and 100°C for two hours in water bath (Memmert, Germany). The antimicrobial compound was also subjected to autoclaving temperature of 121°C for 15 minutes. The effect of pH on the antimicrobial agent stability was also examined. Aliquots of the supernatant were incubated at different pH values, ranging 2-12, at 35°C for 120 minutes in screw capped tubes; then, neutralized to pH = 7.0. Before and after each treatment, the samples were tested for antimicrobial activity against B. subtilis as the indicator organism and an untreated cell-free supernatant served as the control As a control indicating original antimicrobial activity.
3.7. Ammonium Sulfate Precipitation
With the assumption that the antimicrobial compound was a peptide in nature, the ammonium sulfate precipitation was performed. The GA strain was cultured in an optimized basal medium for 72 hours. Then, the culture was centrifuged at 12000 ×g for 10 minutes to remove the cells. Solid ammonium sulfate was added slowly to the culture supernatant to reach 70% saturation and stirred for 24 hours at 4°C. The precipitate was harvested by centrifugation at 18000 ×g for 15 minutes, dissolved in phosphate buffered saline (PBS), and dialyzed against the same buffer overnight at 4°C (cut off 1 kDa). Both the dialyzed sample and the buffer were then assayed for antimicrobial activity.
3.8. Extraction and Polarity Determination of Antimicrobial Compounds
To extract the antimicrobial agent, the culture medium was centrifuged at 8000×g for 10 minutes. The supernatant was filtered with a nitrocellulose membrane filter (pore size: 0.22 µm). The filtrate was mixed with 25% (v/v) of the organic solvents, including chloroform, ethyl acetate, and methanol; then, it was vigorously shaken and allowed to separate. The organic phase was separated from the exhausted supernatant and it was repeated 15 times. The organic and aqueous phases were concentrated under reduced pressure by a vacuum pump at 30°C (R-114, Buchi, Switzerland) until no solvent was left. Then, the two phases were tested for antimicrobial activity by agar disk diffusion method.
3.9. Chromatography and Bioautography of the Antimicrobial Compounds
The methanolic extract of the supernatant was chromatographically analyzed using the thin layer chromatography (TLC) method. The samples were spotted on 2 × 10 cm2 silica gel plates using spotting tubes about 1-2 cm above the bottom of the plates. Then, they were placed in a chromatography tank with selected solvents of different polarities such as hexane, methanol, chloroform, ethanol, distilled water, and mixture of chloroform/methanol (1:3), enough to wet the lower edge of the plate, before the spotting was performed. The plates were left in the solvent for some time, during which, the solvent moved across the plate from bottom to top. The plates were removed from the tank, allowed to dry, and then visualized under ultraviolet irradiations at 254 and 366 nm by spraying with Ninhydrin. The plates with more spots were used in the bioautography test. The silica gel plates were seeded with B. subtilis and incubated for 20 hours at 37°C. The clear zones due to growth inhibition of the microorganisms indicated the location of antimicrobial compounds on the TLC plates.
4. Results
4.1. Identification of the Bacterium
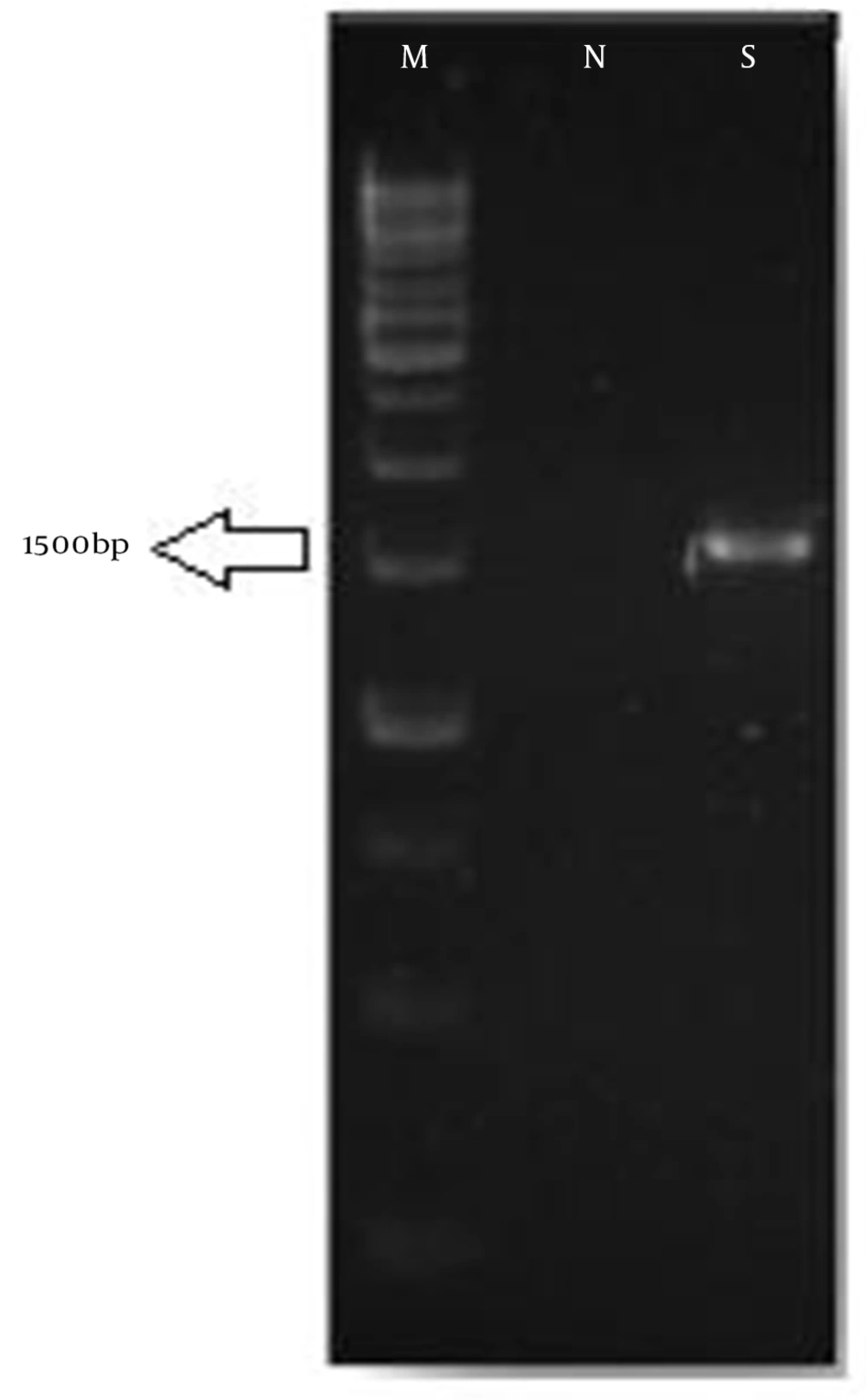
In this study, an aerobic, motile, Gram-positive, catalase-positive, oxidase-negative and spore-forming bacterium was isolated from the frog skin. Morphological and biochemical properties of the isolated bacterium had been investigated by Bergey’s manual of systematic bacteriology (7). Summarized results of the biochemical and morphological tests are shown in Table 1. The sequence of 16SrDNA was obtained after DNA extraction and PCR amplification in 1500 bp (Figure 1). The sequence was reversed, aligned, and compared to similar database sequences, using the Bioedit software Boston, USA. BLAST analysis demonstrated a high level of similarity (95%) to the sequence of B. atrophaeus as well as those of Bacillus sp.
| Characteristic | GA Strain | Bacillus atrophaeus (9) | Bacillus subtilis (7) |
|---|---|---|---|
| Shape | Rods | Rods | Rods |
| Gram staining | + | + | + |
| Motility | + | + | + |
| Aerobiosis | - | - | - |
| H2S production | - | - | - |
| Indole | + | - | - |
| Oxidase | - | - | - |
| Catalase | + | + | + |
| Black pigment production | + | + | - |
| Hemolysis | + | ND | + |
| Urease | - | ND | - |
| Voges-Proskauer test | - | + | + |
| Hydrolysis of gelatin | + | + | + |
| Hydrolysis of starch | + | + | + |
| Glucose | + | + | |
| Lactose | - | - | + |
| Sucrose | + | + | + |
| Mannitol | + | ND | + |
aAbbreviation: ND, not determined.
4.2. Antimicrobial Activity of the GA Strain
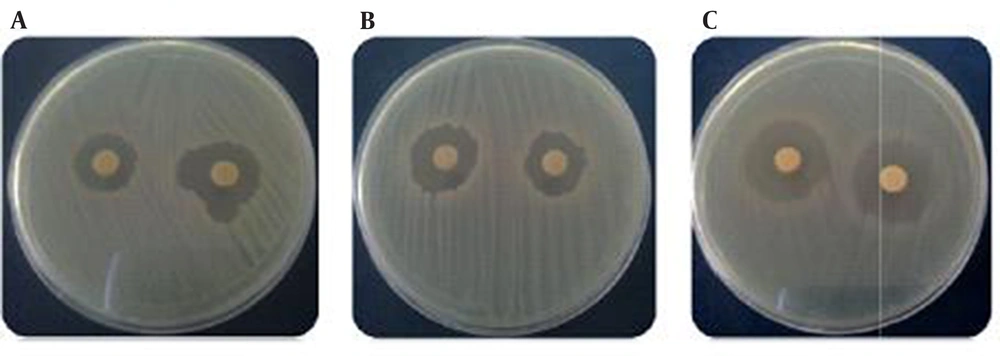
The antimicrobial assay of the GA strain culture supernatant was performed by MIC and disk diffusion methods. The results showed that the GA strain antimicrobial compound was broad spectrum and showed high activity against Gram-positive bacteria, yeasts, and Gram-negative bacteria (Figure 2andTable 2).
| Standard Strain | Zone of Inhibition |
|---|---|
| Bacillussubtilis ATCC 465 | +++ |
| Staphylococcus aureusATCC 25923 | +++ |
| Escherichia coli ATCC 25922 | ++ |
| Pseudomonas aeruginosaATCC 85327 | + |
| Candida albicans ATCC 10231 | +++ |
| Aspergillusniger | ++ |
a Antimicrobial activity of the GA strain against the laboratorial standard bacteria after 18 hours of incubation at 35°C in Muller Hinton agar by disk diffusion method (+ ≤ 10, ++ > 10, +++ ≥ 15 mm).
4.3. Effect of Different Media on Antimicrobials Production
Among different studied media, including nutrient broth, Muller Hinton broth and the synthetic media with glucose and glycerol as carbon source, the maximum antimicrobial production was observed in mineral salt media containing glucose and glycerol. In this media, antimicrobial production started at the late level of logarithmic growth phase.
4.4. Effect of Temperature and pH on Antimicrobial Activity
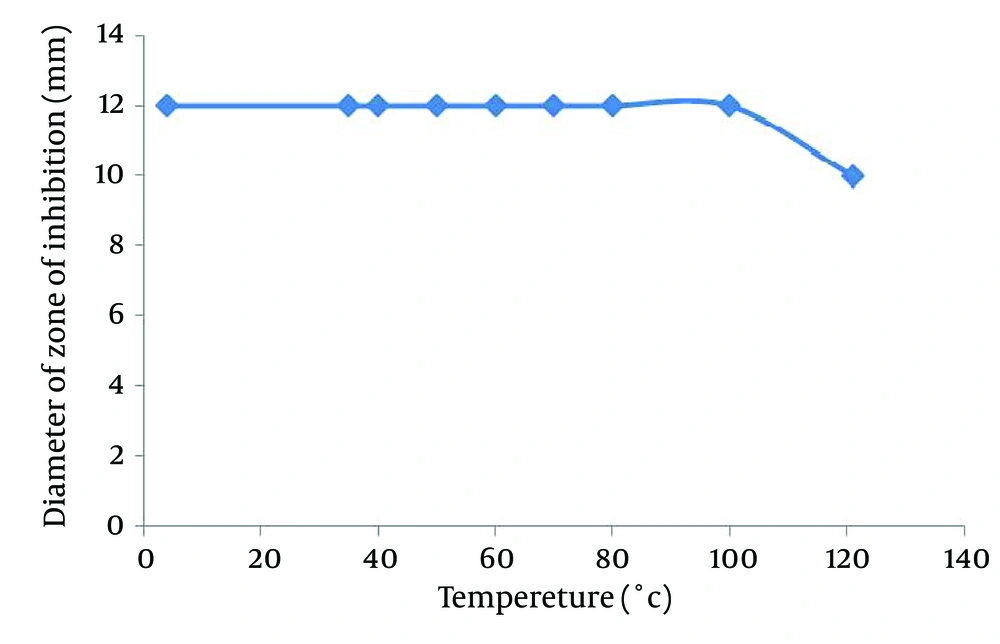
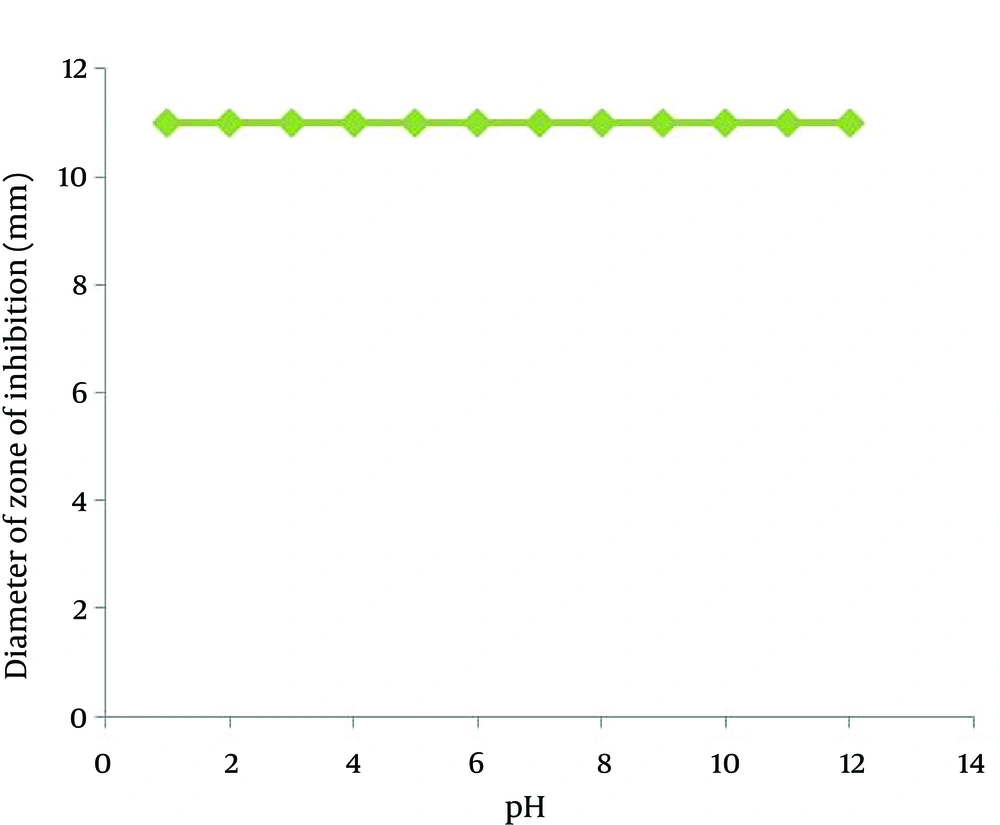
The study on the effect of temperature and pH on antimicrobial activity of the supernatant showed that the supernatant retained its antimicrobial activity when remained at 4, 35, 40, 50, 60, 70, 80 and 100°C for 120 minutes, but autoclaving at 121°C for 15 minutes caused loosing 50% of its antimicrobial activity (Figure 3). The activity of the isolated antimicrobial compound was not affected under a wide pH range from 2.0 to 11.0 (Figure 4).
Aliquots of the culture supernatant were prepared in screw capped tubes, with pH adjusted to 7, and placed in water bath at 4, 35, 40, 50, 60, 70, 80 and 100°C for two hours. The antimicrobial compound was also subjected to 121°C for 15minutes. Before and after the treatment, the samples were tested for antimicrobial activity against B. subtilis as the indicator organism; an untreated cell-free supernatant served as the control negative.
Aliquots of the supernatant were incubated at different pH values ranging 2-12 at 35°C for 120 minutes in screw capped tubes, then neutralized to pH = 7.0. Before and after the treatment, the samples were tested for antimicrobial activity against B. subtilis as the indicator organism and an untreated cell-free supernatant served as the control negative.
4.5. Ammonium Sulfate Precipitation and Molecular Weight Determination
The cell-free supernatant of the bacterium culture was dried under reduced pressure by vacuum pump at 30°C (R-114, Buchi, Switzerland). The dried compounds were dissolved in PBS and dialyzed against the same buffer. The dialyzed samples as well as the outside buffer had antimicrobial activity against B. subtilis. The results showed that molecular weight of the antimicrobial substance was less than 1 kDa. The precipitant of ammonium sulfate had no antimicrobial activity, indicating that the antimicrobial compound did not possess protein properties.
4.6. Extraction of Antimicrobial Compounds
The organic phase and raffinate of the bacterium culture supernatant were separately concentrated and dried on sterile antibiotic assay disks. The inhibition growth zone was observed around disks containing dried extracts of ethylacetate and methanol. Compared to other solvents, chloroform could not recover the antimicrobial agents of supernatant and no antimicrobial activity was observed. The results suggested that the GA strain antimicrobial compound has somewhat amphiphilic properties.
4.7. Isolation of the Active Compound
On the silica gel thin-layer chromatogram, the mixture of chloroform: methanol (1:3 ratio) separated the methanol extract into two bands, which were visualized by Ninhydrin under ultraviolet irradiations at 254 and 365 nm. In the bioautography test, antimicrobial activity of the bands was not observed.
5. Discussion
Synthesis of secondary metabolites by microorganisms, especially antibiotics, is very important in commercial and research point of view (10). Nowadays, indiscriminate usage of antibiotics has resulted in emergence of resistant microorganisms. Therefore, screening researches for products with antimicrobial activities are necessary. The moist surface of amphibians’ skins creates a good place for pathogenic microorganisms (4, 5). One factor that may contribute to disease resistance in the body is innate immune defense, including antimicrobial peptides and symbiotic microorganisms. The symbiotic bacteria on the frog skin surface may be crucial for protection of amphibians against skin-invasive pathogens (6). The GA strain was isolated from the skin of R. ridibunda, a type of frog. According to 16SrDNA sequencing and biochemical and morphological tests, the bacterium was identified as B. atrophaeus. Reports about the antimicrobial compound derived from B. atrophaeus are rare. Jin et al. isolated the B. atrophaeus strain from marine sponge which produced Bacillamide C with only anti algae activity (11).
Most of the species from the genus Bacillus are considered as industrially important bacteriocin producers and have a history of safe usage. Bacteriocins are peptides and protein antibiotics produced by several species and have antimicrobial properties usually against other closely related species (12). The results of this study showed that antimicrobial substance from the GA strain differed from antibiotics produced by bacteria of the genus Bacillus in some properties. Unlike bacteriocins, the GA strain antimicrobial compound was broad spectrum. According to Abada et al. broad-spectrum antibiotics are compounds that have activity against Gram-negative and Gram-positive bacteria as well as against fungi (13). The GA antimicrobial compound showed high activity against Gram-positive bacteria (S. aureus), yeasts (C. albicans) and Gram-negative bacteria (E. coli, P. aeruginosa).
Maximum production of the antimicrobial compound was achieved at 30ºC and pH = 7.0. The bacterium was also capable of producing antimicrobial agents even at low temperatures (25ºC). Carbon source is an important part of medium for bacterium growth and antimicrobial production. The results showed that glucose and glycerol were good sources for growth and high production of antimicrobial agents. The GA strain antimicrobial substance exhibited heat stability between 25°C and 100°C for 120 minutes and autoclaving at 121°C for 15 minutes caused loss of 50% of its antimicrobial activity. A staphylococcal antimicrobial peptide, produced by S. hominis KQU-131, was found to lose 50-75% of its antimicrobial activity when heated at 100°C for 100 minutes and at 121°C for 15 minutes. Kim et al. described the biochemical and structural properties of the antimicrobial peptide produced by S. hominis MBBL 2-9 (14). This peptide antibiotic, namely hominicin, showed heat stability up to 121ºC for 15minutes. They suggested that the thermal stability of hominicin comes from its structural properties. Therefore, same as hominicin, the thermal stability of the GA strain antimicrobial compound may be due to its small size and structural limitation.
The isolated antimicrobial compound was stable and active under a wide pH range from 2.0 to 11.0. The hominicin, produced by S. hominis MBBL 2-9, showed activity under both acidic and basic conditions, pH range of 2.0-10.0 (14). Kumar et al. reported peptide antifungal antibiotics driven from B. subtilis (15). Same as the GA antimicrobial agent, this peptide antibiotic was active in a broad range of pH, but the methanolic extract of this compound had no antimicrobial activity. Other recently reported antimicrobial peptides not only lose their activities under extreme acidic conditions, but also have a gradual decrease in antimicrobial activities at basic pH levels. Therefore, these antimicrobial peptides have a considerable disadvantage to be used as food additives, because many foods have a wide pH range from extreme acidic to basic. By contrast, the GA antimicrobial substance showed adequate stability at a wide pH range between 2.0 and 11.0.
In this study, an antibiotic-producing bacterium that was isolated from the frog skin of R. ridibunda, was identified as B. atrophaeus. The antimicrobial compound produced by the bacterium was broad-spectrum antibiotic, exhibited heat stability, and was active under both acidic and basic conditions. These properties make the compound as a valuable tool not only for prevention and treatment of infections, but to be considered as a preservative in food industries to avoid food deterioration and spoilage. However, further studies on purification and characterization of the intended antibiotic would be inevitable.