1. Background
The difficulties seen in combating infections caused by drug resistant organisms, especially those that have acquired resistance to beta-lactams, such as broad-spectrum cephalosporins, have resulted from the extensive use of broad-spectrum antibacterial agents. One important resistance mechanism to the beta-lactams, including new cephalosporins, is due to the destruction of the antibiotics by extended-spectrum beta-lactamases (ESBLs) (1). ESBLs were first described in the early 1980s among Klebsiella species, and then in Escherichia coli, Serratia marcescens, Pseudomonas aeruginosa, and other Gram-negative bacilli (2, 3). ESBLs are a group of enzymes that have an expanded substrate profile which allows for the hydrolysis of 3 and 4 generation cephalosporins and monobactams, but not carbapenems. ESBLs are prevented by β-lactamase inhibitors, such as; clavulanic acid, sulbactam and tazobactam (2, 4). These enzymes can be either plasmid or chromosomally mediated, but they are described mainly on plasmid that are frequently found among Enterobacteriaceae.
ESBLs are prevalent in every part of the world, and in addition, they are found in a meaningful percentage of E. coli and Klebsiella pneumoniae strains in certain countries (4, 5). ESBLs are continuous mutations that change the amino acid configuration near the active site of these β-lactamases, resulting in the development of new enzymes showing extended substrate profiles. Until now, more than 400 different ESBLs have been identified, and these are clustered into three groups: TEM, SHV and CTX-M, with 183, 134 and 103 variants, respectively. Among the previously mentioned ESBL variants, TEM and sulphydryl variable SHV were the major types in some countries (6-8).
Determination of ESBL genes, including TEM and SHV, by molecular techniques in bacteria that produce ESBL and their antimicrobial resistance patterns can provide applicable information about their epidemiology and risk factors related to their infections (2, 9). A number of studies have been carried out to recognize the types of ESBL producing Enterobacteriaceae in hospitals in Iran (1, 10, 11). In spite of the presence of ESBLs among K. pneumoniae and E. coli which have been reported from Isfahan (12), there is no information on their molecular types.
2. Objectives
The present study was conducted to determine the prevalence of TEM and SHV genes responsible for ESBL production amongst ESBL positive K. pneumoniae and E. coli isolated from urinary tract infection specimens of both hospitalized patients and outpatients.
3. Materials and Methods
3.1. Bacterial Isolates
In this study, E. coli and K. pneumonia strains isolated from patients suffering from urinary tract infections were studied. The isolates were collected from December 2011 to October 2012 from Al-Zahra Hospital, Isfahan. Tests were conducted on both hospitalized and non-hospitalized infections. Hospitalized infections were defined as patients who were confined to bed in hospital, while non-hospitalized infections were defined as infections in patients who had had no previous contact with hospitals or long-term care facilities in the previous two weeks. Bacterial isolates were characterized using biochemical tests. The samples were cultured on nutrient agar (Hi Media, India), MacConkey agar (Hi Media, India), blood agar (Hi Media, India) and eosin methylene blue (EMB) agar (Hi Media, India). The plates were incubated at 35°C for 24 h and the pure isolates characterized and identified according to Gram stains and biochemical tests such as; catalase, oxidative, indole production, citrate utilization, triple iron sugar, urea test, oxidative-fermentative test with glucose, ortho-nitrophenyl-β-galactoside (ONPG) test, and methyl red Voges-Proskauer, as described in standard bacteriological methods. All of the above chemicals and media were purchased from Sigma-Aldrich (Germany).
3.2. Antimicrobial Drug Susceptibility Testing
Antimicrobial susceptibility of the isolated bacteria was performed by the disk diffusion technique on Mueller-Hinton agar plates (Oxoid, UK), according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI). The antibiotics (µg) tested included: amikacin (30), ampicillin (10), ciprofloxacin (5), co-trimoxazole (10), gentamicin (10), imipenem (10), nitrofurantoin (300), tazocin (110), ceftazidime (30), cefepime (30), nalidixic acid (30), and cefotaxime (30). The standard antibiotic disks were purchased from Mast Diagnostics (Mast Group, UK).
3.3. Phenotypic Screening of Extended-Spectrum β-lactamase
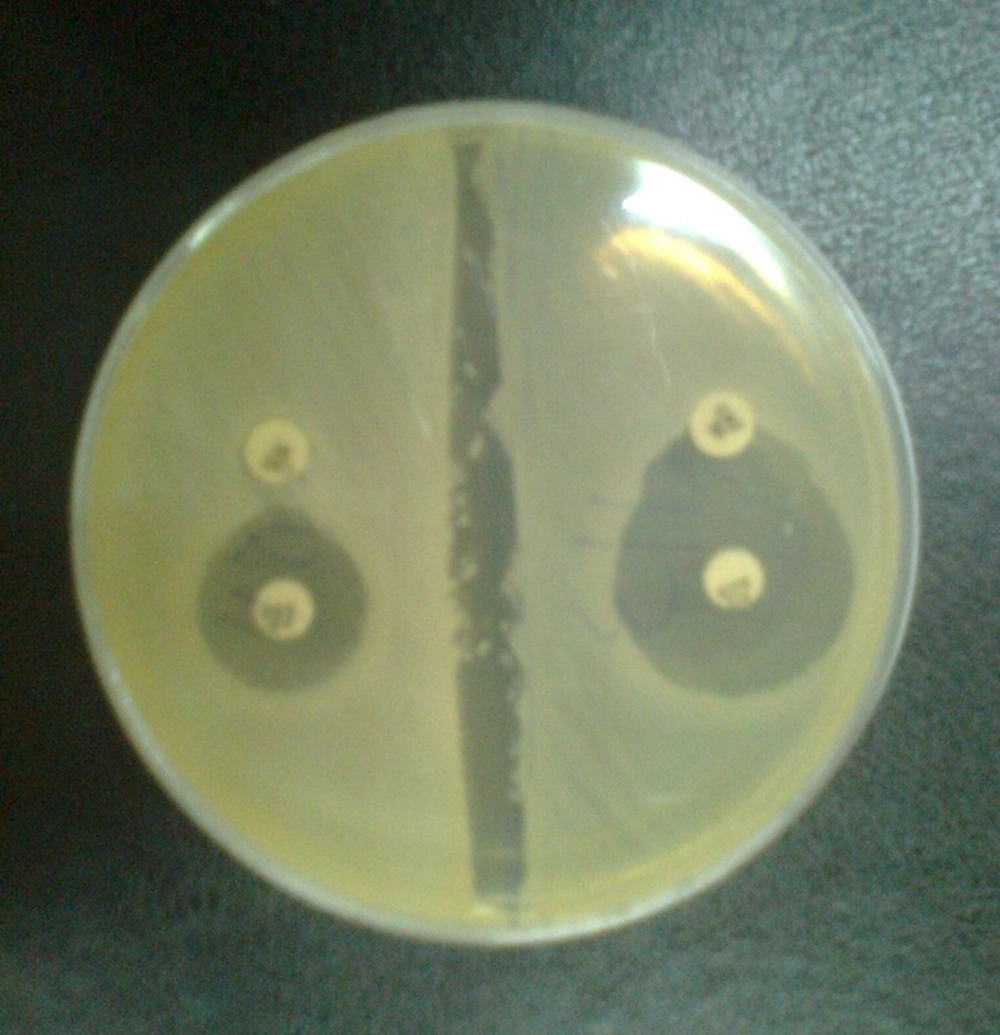
The isolates that showing resistance to one or more third generation cephalosporins (3GCs) were examined for ESBLs production by the combination disc method using; cefotaxime (30), cefotaxime/clavulanic acid (30/10), ceftazidime (30), and ceftazidime/clavulanic acid (30/10) (MAST Co. UK). A greater than or equal to 5mm increase in diameter of the inhibition zone of the cephalosporin-plus-clavulanate disc, when compared to the cephalosporin only disc, was interpreted as phenotypic evidence of ESBLs production. K. pneumoniae ATCC 700603 was used as a positive control and E. coli ATCC 25922 was used as a negative control. Standard strains were obtained from the American Type Culture Collection (Manassas VA).
3.4. DNA Extraction for Polymerase Chain Reaction
DNA templates for polymerase chain reaction (PCR) were obtained from the overnight growth of bacterial isolates on Luria–Bertani agar (Hi Media, India) that were pelleted by centrifugation, resuspended in 500 µL of sterile deionized water, and then boiled for 10 min. After centrifugation of the boiled samples at 19000 g for 5 min, the upper layer was used as a DNA template for PCR.
3.5. Polymerase Chain Reaction Amplification of β-Lactamase Genes
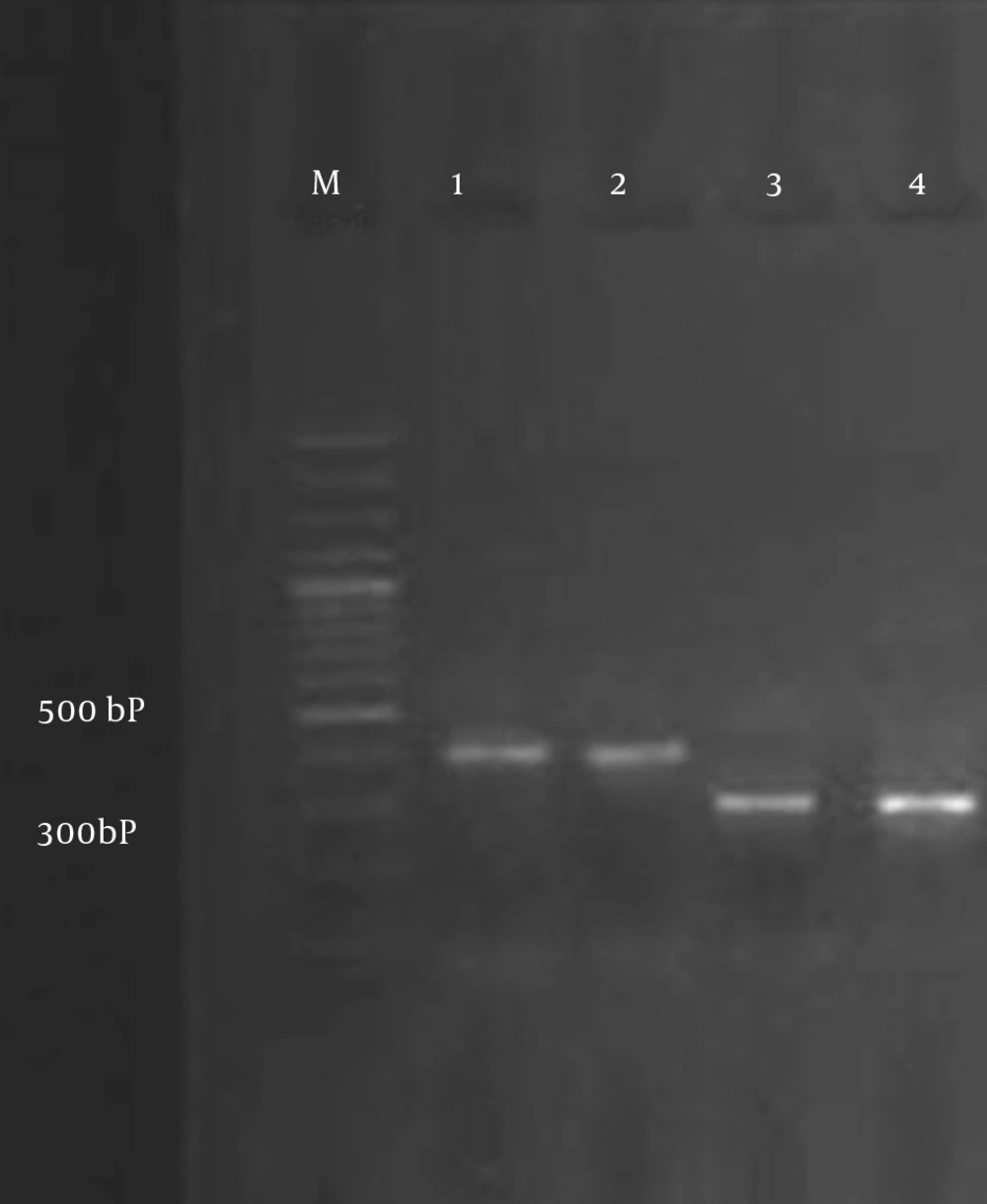
Bacterial isolates included in the study were screened by PCR for TEM and SHV genes. Specific primers and annealing temperature for amplifying the blaSHV and blaTEM genes by PCR are shown in Table 1. PCR was carried out in a 25 µL volume containing; 2.5 µL of 10X PCR reaction buffer, with MgCl2 (1.5 mM), 0.5 µL (200 µM) deoxynucleoside triphosphate mix (dNTPs, 10 mM), 0.5 µL of each primer (10 pm/µL), and 50 ng DNA template with 0.5 µL (3 U/µL) Taq DNA polymerase. PCR amplifications were performed on a Thermocycler TC-512 (Techne) and the PCR fragment was analyzed on 1% agarose gel. A molecular marker (Fermentase SM024: 80 to 1000 kb) was used to assess the PCR product size. PCR materials were purchased from Fermentase (Life Science, Germany).
aAbbreviations: SHV, Sulfhydryl Variable; F, Forward; TEM, Temoneira; R, Reverse.
4. Results
During a 10-month period, 245 E. coli and 55 K. pneumonia strains were isolated from patients with urinary tract infections. In total, 128 of the 300 isolates were confirmed as potential ESBL producers using ceftazidime/clavulanic acid and cefotaxime/clavulanic acid disks (Figure 1).
The occurrence of ESBL in the isolates was as follows: 107 (43.67%) out of 245 E. coli and 21 (38.18%) out of 55 K. pneumonia. Of the 107 E. coli, 67 (62.62%) were isolated from hospitalized patients and the remaining 40 (37.38%) belonged to outpatients. Furthermore, of the 21 K. pneumonia, 13 (62%) were from hospitalized patients, while 8 (38%) were recovered from outpatients.
All ESBL-producing isolates were screened by PCR using blaTEM and blaSHV specific primers (Figure 2). ESBL genes were found in 24 isolates (18.75%): 21 E. coli and 3 K. pneumonia isolates. The overall data revealed that the TEM gene was present in 13 (12.14%) E. coli, but it was not detected in K. pneumonia, and the SHV gene was present in 8 (7.47%) E. coli and 3 (14.28%) K. pneumonia isolates. There were 5 (4.67%) E. coli isolates that harbored both TEM and SHV genes. All bacterial isolates were susceptible to imipenem. The lowest rates of resistance to other antibiotics were observed for: tazocin (6.25%), amikacin (12.5%) and gentamicin (14.84%). The rates of resistance to other antibiotics were as follows: nitrofurantoin (16.4%), nalidixic acid (23.43), co-trimoxazole (25%), cefepime (32%), ciprofloxacin (55.46%), ceftazidime (59.76%), ampicillin (69.53%) and cefotaxime (73.43%).
5. Discussion
The treatment of infectious diseases is an important issue for human wellbeing and the daily increase in bacterial resistance has elevated patients’ costs in recent years. The production of ESBLs is also a major threat to the use of the new generation of cephalosporins. In the last two decades, the rate of ESBL production by Enterobacteriaceae has increased considerably (10, 13, 14). Among Enterobacteriaceae, K. pneumonia and E. coli are the most important causative agents of nosocomial infections (15) and these are usually isolated from patients with urinary tract infections in Al-Zahra Hospital, Isfahan; therefore, we selected these two types of bacteria for our study. Occurrences of infection effected by extended spectrum beta-lactamase producing K. pneumonia and E. coli have been widely reported all over the world following the widespread use of the expanded spectrum cephalosporins (11, 16-19).
In our study, phenotypic screening of ESBL showed that 43.67% of E. coli and 38.18% of K. pneumonia isolates were positive for ESBL production. Overall, 62.62% of E. coli were isolated from hospitalized patients and the remainder 37.38% belonged to outpatients. In addition, 62% of K. pneumonia were from hospitalized patients, while 38% were recovered from outpatients. Based on these results, the prevalence of ESBL producing K. pneumonia and E. coli was high. In addition, ESBLs producing organisms were higher in hospitalized patients compared to outpatients. The high prevalence of ESBLs producing E. coli and K. pneumonia has also been reported by a number of previous studies. For example in a study by Feizabadi et al. (1), they showed a 44.5% ESBL positive rate among clinical K. pneumoniae isolated from clinical specimens in Tehran (20).
In a study from Kurdistan, Ramazanzadeh et al. revealed a 34.8% ESBL positive rate among strains of Gram-negative bacteria (21) and Mobasherizadeh et al. showed that among a total of 2035 consecutive clinical isolates identified as E. coli in Al-Zahra Hospital, 898 (44.1%) and 432 (21.2%), were ESBL producers for hospitalized and non-hospitalized patients, respectively (22). Therefore, our findings were in agreement with the above-mentioned studies. On the other hand, the rate of ESBLs production is higher in other hospitals or cities in our country. For instance, the rates of ESBL producing K. pneumoniae isolated from Tehran indicated by Aminzadeh et al. were 52.5% in 2008 (23). Bazzaz et al. also showed that the prevalence of ESBL positive strains of E. coli and K. pneumonia was 59.2% in 2009 (24).
In another study that was performed by Jalalpoor and Mobasherizadeh. in 2009-2010 in Isfahan (12), the frequency of ESBLs in strains of E. coli in hospitalized and out-patients was 58% and 17%, respectively, and the frequency of ESBLs in strains of K. pneumoniae in hospitalized and out-patients was 64% and 22%, respectively. In that study, the samples were collected from Al-Zahra, Shariati, and Kashani hospitals and reference and Mahdieh laboratories. Their study results showed a higher frequency of ESBLs in isolated bacteria from hospitalized cases when compared to out-patients. Therefore, we see that our results are compatible with the results of the previously mentioned study. The presence of ESBL positive Enterobacteriaceae was also documented by Feizabadi et al. (1). From 104 isolates of K. pneumonia collected from four university hospitals in Tehran, they identified 75 (72.1%) ESBL producing isolates. Moreover, E. coli and K. pneumonia isolates producing ESBL are less prevalent in the Al-Zahra Hospital, Isfahan. In a study from Mashhad, Zaniani et al. revealed that 43.9% of E. coli and 56.1% of K. pneumoniae were ESBL producers and the frequency of SHV and TEM among the ESBL producing isolates were 14.4% and 20.6%, respectively (25). The prevalence of ESBLs varies from one hospital to another. It is uncertain whether this is because of the differences in infection control practices between hospitals or to differences in the use of new cephalosporins (11).
Our study, along with other studies, have also demonstrated that the rates of ESBLs production in our country are different from other countries in our area, such as; India (57.1%), Turkey (57%) and South Korea (30%), and published data from European countries, such as; France, Italy, the Netherlands, Germany, and Spain, as well as in the United States, Australia, Japan, Tanzania, Thailand and Pakistan, which showed a higher prevalence of ESBL-producing isolates in the present study (22, 26-28). Feizabadi et al. (1) found that the rates of resistance for tazocin, amikacin, ciprofloxacin, cefepime, ceftazidime, and cefotaxime were; 21.3%, 21.4%, 28%, 76% and 84.0%, respectively. The comparison of our study results with the above-mentioned study shows that antibiotic resistance to four of the previously mentioned antibiotics is lower in our study. In addition, in another study, both non-hospitalized and hospitalized isolates were more resistant to first line drugs including; ampicillin, and trimethoprim-sulfamethoxazole (22). This result, which is comparable with other studies in developing countries, is due to the widespread use of these drugs because of their low cost and easy administration.
Imipenem, amikacin and piperacillin-tazobactam were the most effective antibiotics against hospitalized ESBL-producing isolates. All ESBL-positive isolates were susceptible to imipenem, indicating that this agent is the best drug for treating serious infections caused by ESBL-producing E. coli. The carbapenem antibiotics including; ertapenem, imipenem, and meropenem, are commonly known as the first choice in the treatment of serious infections caused by ESBL-producing Enterobacteriaceae (22). The prevalence of the TEM gene in our study in E. coli was 12.14%, however, this gene was not detected in K. pneumonia. The prevalence of the SHV gene in E. coli and K. pneumonia isolates was 7.47% and 14.28%, respectively, which is lower than that reported by Karimi et al. (26% and 15% for E. coli, and 18% and 15% for K. pneumonia, respectively) (15), 54% and 67.4% for K. pneumonia (29) and 55.7% and 30.7% for K. pneumonia (1). Also in this study, 5 (4.67%) of the E. coli isolates harbored both TEM and SHV genes which is lower than that reported in India (67.3% 2003 to 2004) (26), (21.8% 2007 to 2008) in Kashan, Iran (10) and (34.7% 2006 to 2007) in Tehran, Iran (1). Therefore, we can conclude that the production of β-lactamase can result from other types of ESBLs among isolated Enterobacteriaceae. The emergence and expansion of ESBL-producing Enterobacteriaceae including, E. coli and K. pneumoniae is an alarming issue and the use of cephalosporins against these isolates is inefficient.
Nowadays, carbapenems are the drug of choice for the treatment of serious infection diseases in Iran. However, the appearance of resistance in this group of antibiotics, especially resistance to imipenem, may restrict their prescription in the future. Another concern is the tremendous increase in the prevalence of ESBLs, as the majority of these were resistant to other antibiotics. Because of the significance of ESBL producing Enterobacteriaceae including, E. coli and K. pneumoniae and the difficulties involved with the therapy of infections caused by these bacteria, clinical laboratories should adopt the employment of a simple test on the basis of the Clinical and Laboratory Standards Institute (CLSI) suggestions for the rapid identification and confirmation of ESBL production in Enterobacteriaceae. In addition, beta-lactam antibiotics and beta-lactamase inhibitors or carbapenems should only be prescribed based on an antibacterial susceptibility test.