1. Background
Influenza A viruses belong to the Orthomyxoviridae family can infect a wide range of birds and mammals including human beings. Viral genome consisting of segmented RNA could result in genetic reassortment between human and animal viruses leading to a pandemic Influenza (flu). Annual epidemic and local outbreaks occur because of highly variable mutational antigenicity in viral surface glycoproteins, hemagglutinin and neuraminidase. Hence no real long-lasting protective vaccine has been developed yet. The current licensed inactivated influenza vaccineis prepared annually based on the WHO recommendation to match the vaccine strain and circulating virus (1, 2). On the other hand, emergence of new strains of influenza virus is unpredictable, as evidenced by the first 21 th century pandemic flu. Therefore, the necessity of preparing an improved universal vaccine using conserved viral proteins is highlighted. It is expected that the vaccine provides broad protection against various viral subtypes and acts as a first line of host protection (3, 4).
Several studies confirmed that DNA vaccines, encoding variable and/or conserved protein of influenza viruses might be effective in animal models as well as humans (5, 6). Vaccination with recombinant DNA could induce accelerating immune responses against conserved epitopes of influenza A viruses and eliminate the risk of using live virus vaccines. The viral ion channel M2 encoded by segment 7 of influenza virus genome is highly conserved among different influenza A subtypes. The protein is expressed in plasma membrane of infected cells and responsible for protein translocation. It has been revealed that DNA vaccines encoding the extracellular domain of the M2 protein (M2e) potentially elicits broad spectrum immunity against homologous and heterologous influenza viruses. Despite very low and even undetectable anti-M2 immune responses in natural infection, different studies have shown that M2 antibodies recognize ectodomain of the protein could reduce the size of viral plaque in vitro, probably through its direct effect on the virus (7, 8). Furthermore, passive transfer of anti-M2 antibody protected mice against fatal viral challenge suggests an antibody-dependent cellular cytotoxic activity.
It could reduce viral replication in vivo to diminish the influenza disease (9). Since 1990 when DNA vaccine introduced, several researches have been performed to improve the vaccine potency by co-administration of chemokine and growth factors or using different adjuvants. Among them, fusing of vaccine antigen to a molecular adjuvant like heat shock proteins (HSPs) and CPG motif has been investigated widely (10, 11). HSPs are highly conserved intracellular chaperons and have been found in both mammalian cells and microorganisms (12). Some of them are potent inducers of immunity and have been used as vaccine adjuvants targeting cancers and infections (13). The action of HSPs as the second signal could stimulate antigen- presenting cells to induce proinflammatory cytokines and promote TH1 responses. In addition, HSPs stimulate upregulation of costimulatory molecules required for priming naive T cells. Therefore, the independent immunostimulatory capacity of HSPs makes them an attractive reagent to induce specific immune responses (14-16).
2. Objectives
The immunostimulatory effect of Leishmania major HSP70 on humoral and cellular responses was well studied in a previous study (17). In the present study, we developed a chimerical DNA vaccine encoding M2 and L. major HSP70 (amino acids 221-604) and evaluated its expression in eukaryotic cells.
3. Materials and Methods
3.1. Cells and Bacterial Hosts
COS-7 cell line purchased from the National Cell Bank of Iran (NCBI) was used for protein expression. The cells were cultured in DMEM (Gibco, Germany) supplemented with 10% FBS and antibiotic solution including 100 U/mL penicillin and 100 µg/mL streptomycin. TOP10F' Escherichia coli was used for cloning experiments and plasmid preparation.
3.2. Cloning of M2 Segment into pGEM-T Easy Vector
A specific primer set previously used for amplification of M2 DNA (18) was modified to construct the chimer gene. In case, the termination codon at reverse primer was removed and a Kozak sequence was added in forward primer to enhance initiation site of translation. The PCR product was run on 2% agarose gel electrophoresis and the distinct band (300 bp) extracted using Qiagen gel extraction kit (Qiagen, Germany) according to the manufacturer's instructions. The purified gene was ligated to pGEM-T easy vector using T4 DNA ligase (Promega, Germany) and transformed into Top10F’chemically competent E. coli cells. Blue-white screening bacterial colonies were performed to isolate the resembling positive colonies, confirmed by PCR and restriction enzyme analysis. The accuracy of cloning was confirmed by sequencing of recombinant plasmid and named pGEM-M2 encoding M2 protein without stop codon.
3.3. Construction of Chimera Expression Vector
Recombinant pGEM II containing L. major HSP70 (nt 661-1812) constructed previously (17), was digested with BamHI and Hind III (Fermentas, Canada). HSP70 DNA gel purified fragment annealed by ligation to the same restriction enzymes digested pcDNATM3.1/Hygro (-) expression vector (Invitrogen, USA), downstream of the CMV promoter and transformed into Top10F’ cells. The confirmed construct named pcDNA-HSP70. The digested M2 DNA fragment obtained from pGEM-M2 was subcloned in the corresponding sites of the pcDNA-HSP70 upstream of the HSP gene. The resultant plasmid named pcDNA-chimer after verification.
3.4. Sequence Analysis and Prediction of 3D Structure
The amino acid sequence of M2 protein, HSP70 and chimer protein were analyzed using web based B-cell epitope prediction algorithms; Bcepred http://www.imtech.res.in/raghava/bcepred and http://www.imtech.res.in/raghava/cbtope for continuous and discontinuous B-cell epitope prediction, respectively. The tertiary structure of proteins was analyzed using online software, Swiss-PdbViewer and WebLab Viewer. Furthermore, SCRATCH servers available at http://www.igb.uci.edu/ were used for protein structure prediction by PSI-BLAST and neural networks.
3.5. Eukaryotic cells Transfection
To express recombinant proteins in eukaryotic cells, pcDNA-M2 (17) and pcDNA-chimer were transfected into eukaryotic COS-7 cells. The cells were seeded in 6-well plates at a density of 6×104/well and transfected with recombinant vectors using Lipofectamine 2000 reagent (Invitrogen, USA) according to the manufacturer instructions. The uninfected cell was considered as control. Efficiency of the transfection procedure was evaluated by transforming EGFP plasmid in parallel. The cell cultures were monitored microscopically two days after transfection.
3.6. RT-PCR for Confirming Expression of M2-HSP70
To determine the presence of M2-HSP70 mRNA in the Cos7 transfected cells, RT-PCR was performed after 48 hours of transfection. Total cellular RNA from transfected and non-transfected cells were extracted and the contaminating DNA was removed by DNase (Fermentas, Lithuania) treatment. The extracted RNA was used in RT-PCR using M2 specific primers.
3.7. Determination of Recombinant Protein Expression by Immunofluorescence Staining
The expression of M2 and chimer proteins was determined by immunoassay of transfected cells using FITC-labeled antibody. Briefly, transfected COS-7 cells grown on glass slides were fixed with cold acetone/ethanol (1:1) for 15 minutes, rinsed with phosphate-buffered saline (PBS) and incubated with 14C2 mouse monoclonal anti-influenza M2 antibody (Abcam, UK) diluted 1:500 in PBS for 40 minutes at 37˚C. The cells rinsed again and treated with FITC-labeled goat anti-mouse IgG (Dako, Denmark) for 30 minutes at 37˚C. Following rinsing with PBS, the slides were visualized by fluorescence microscopy.
3.8. Verification of M2 and Chimer Protein Expression by Western-Blotting
Protein expression was verified by immunoblotting using specific monoclonal antibody as described previously (18). In brief, after 48 hours of transfection, the cells were washed with PBS to remove FBS residues, scrapped into Tris buffer (pH 8) then frozen and thawed three times followed by centrifugation for 15 minutes at 13000 rpm (4˚C). The cell lysates were mixed with an equal volume of 2xSDS sample buffer (4% SDS, 20% glycerol, 125 mM Tris-HCl pH 6.8, 10% 2-mercaptoethanol and 0.01% Bromophenol Blue) and boiled for 10 minutes. Protein samples were run on 15% SDS-PAGE gel with an approximate concentration of 20-40 μg followed by transferring onto nitrocellulose membrane. Subsequently, membrane was probed by 14C2 mouse monoclonal antibody and HRP-conjugated anti-mouse IgG (Sigma, UK) as primary and secondary antibodies, respectively. The protein bounds were visualized by staining the membranes with TMB (Sigma, UK).
4. Results
4.1. Construction of Recombinant Vector
Influenza A virus M2 and L. major HSP70 genes were joint together at their overlapping BamHI restriction site to form a chimer gene, which encoded M2 and HSP70 protein in a single ORF. Arrangement of fragment junctions is shown in Figure 1.
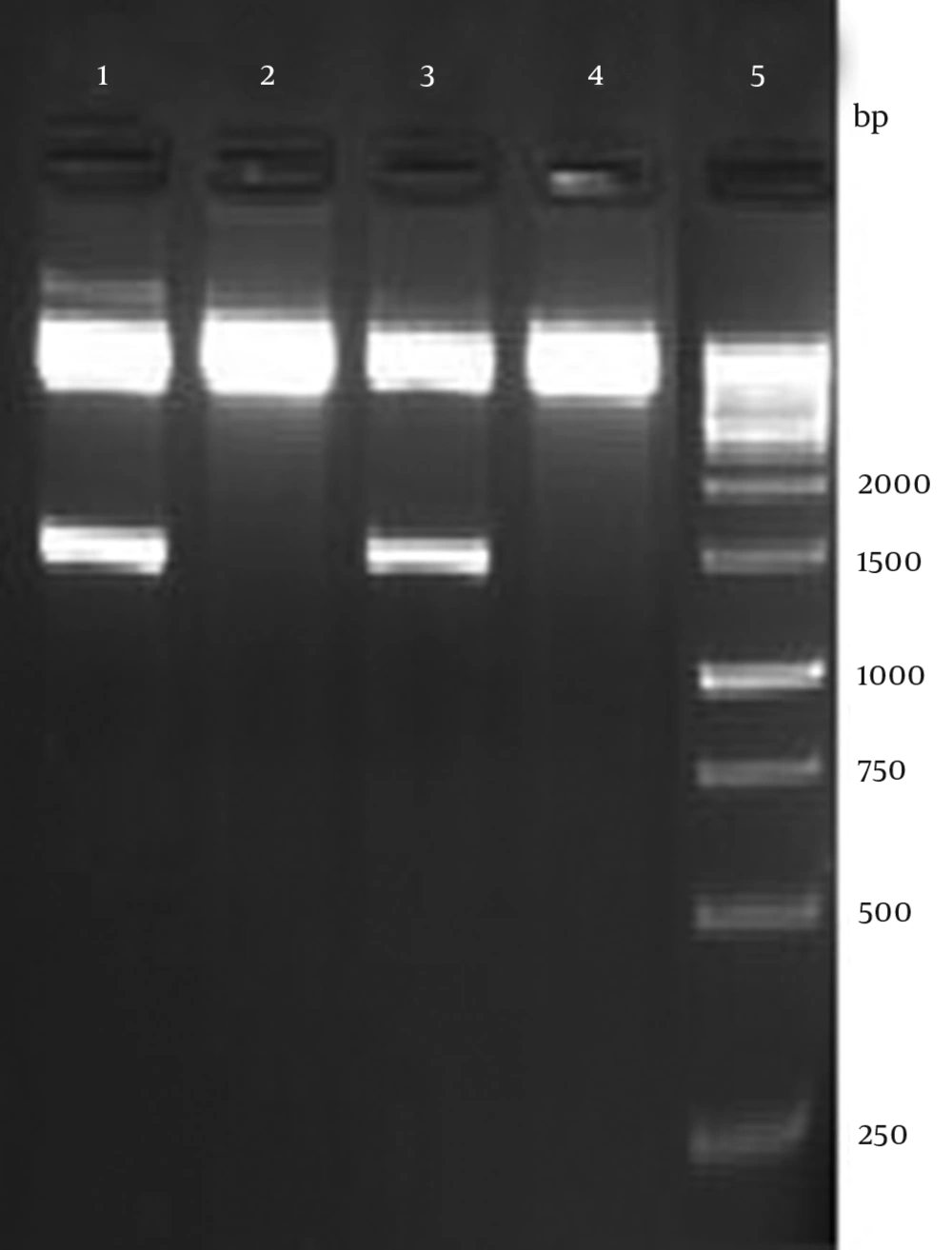
The recombinant plasmid was verified by enzymatic digestion with HindIII and XhoI resulted in 1452 bp fragment as expected (Figure 2). Accuracy of the cloning procedure was confirmed by sequencing of recombinant plasmid by Eurofins MWG, Germany. Analysis of sequencing was accomplished using Chromas software (version 1.45 - Australia).
4.2. Prediction of 3D Structure
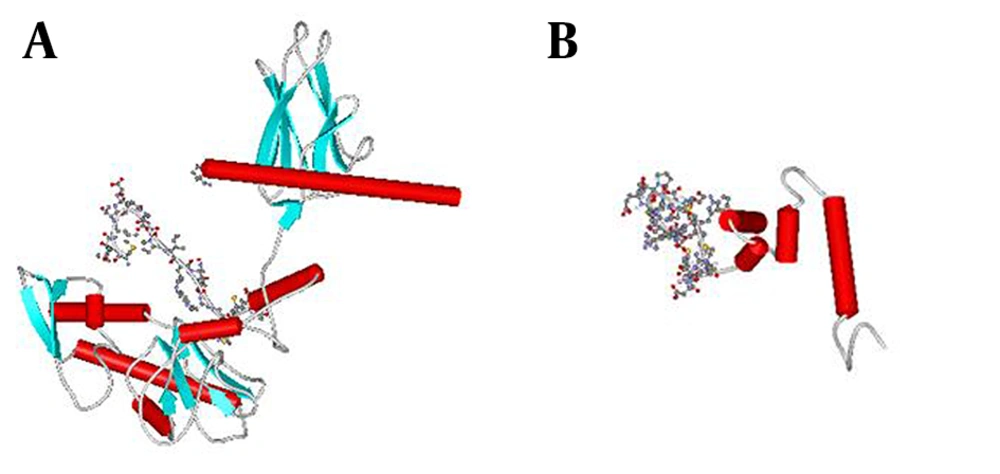
To predict and analyze the tertiary structure of recombinant proteins, we used Swiss-Pdb Viewer, WebLab Viewer and SCRATCH. The prediction result showed that fusing HSP to the C- terminal of M2 protein does not mask the predominant epitope (M2e) as shown in Figure 3.
4.3. Detection of Specific mRNA
Transcription experiment in transfected cells was confirmed by PCR, which was performed on cDNA by the M2 specific primers (data not shown).
4.4. Antigenicity Assessment of the M2 and M2-HSP70 Proteins
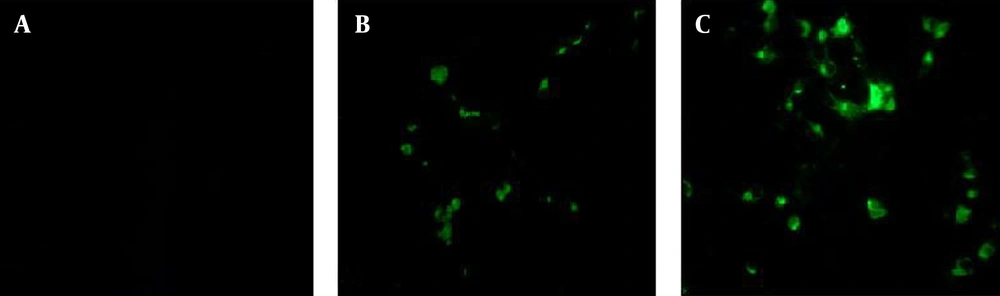
Immunofluorescence was performed to evaluate the antigenic potency of the recombinant proteins. As illustrated in Figure 4, the fluorescence emission was detected in cells transfected with recombinant plasmids containing M2 and M2-HSP70 genes; whereas, uninfected cells did not represent any fluorescence emission.
4.5. Expression and Confirmation of the Recombinant M2 and Chimer Protein
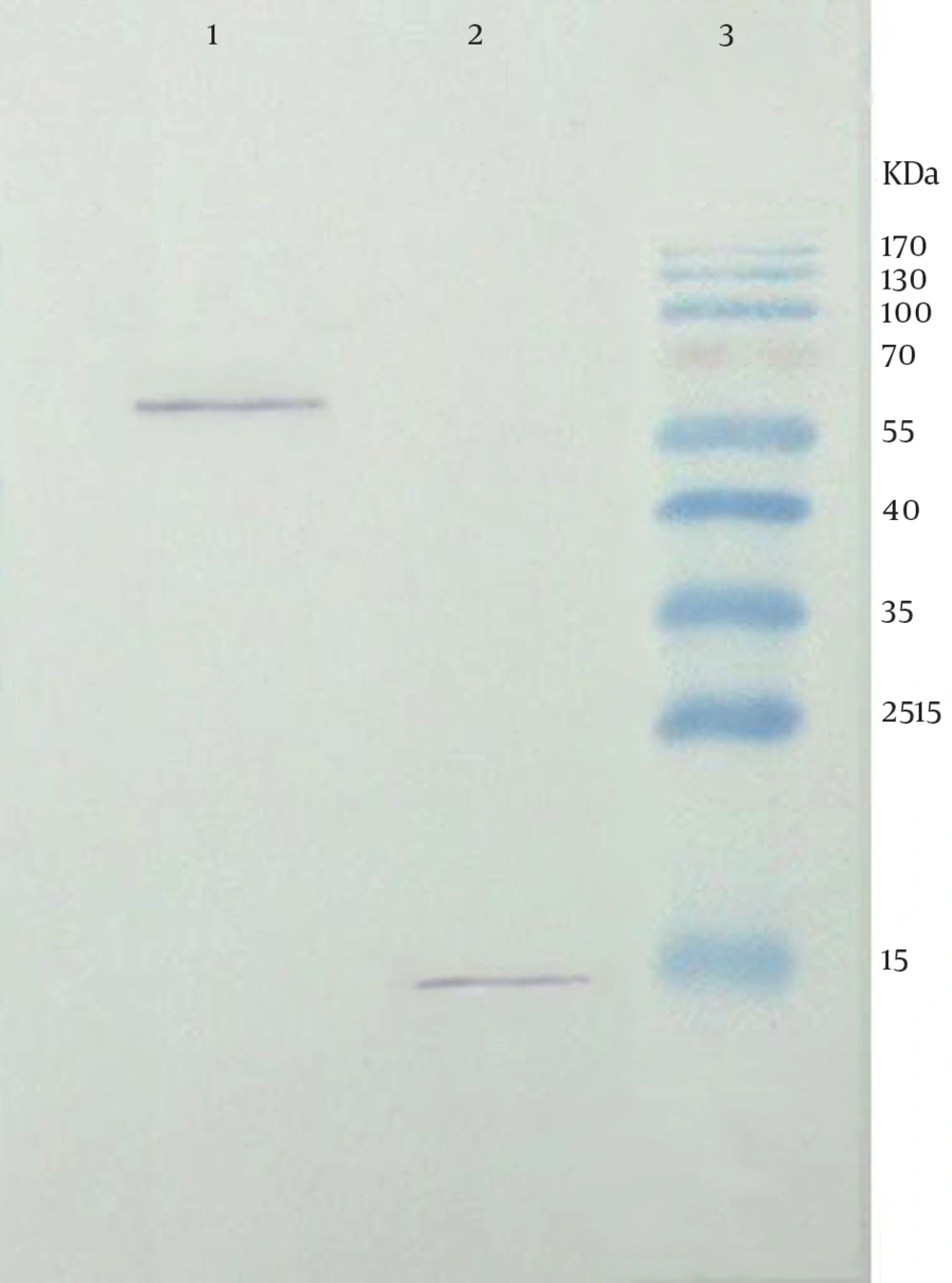
Confirmation of the recombinant fusion protein was performed by blotting of the transfected cell lysate. Following treatment with HRP-conjugated anti-mouse IgG, the expected bands at approximate weight of 15 and 60 kDa were observed related to M2 and chimer protein, respectively (Figure 5).
5. Discussion
Protective immunity against influenza virus is mainly induced by serum antibodies. The most important antigenic component of the commercial vaccines is viral hemagglutinin which is frequently mutated leading to antigenic variation. The variability in the major neutralizing antigens allows the virus to escape from preexisting anti-influenza host immune system. It is necessary to update the vaccine strains considering recently circulating virus each year to produce a protective vaccine (19). Moreover, in the case of an unexpected pandemic flu, it takes a long time to monitor a new strain to produce a well-match vaccine in industrial scale. Developing an effective universal vaccine using conserved viral proteins with low antigenicity, which provides a comprehensive protection against different influenza viruses is more promising to control a threatening pandemic and could save time to produce a protective vaccine.
The effectiveness of M2 protein as a universal vaccine has been demonstrated extensively. Slepushkin et al. (20) found that full length M2 protein expressed in insect cells induces a heterosubtypic protective immunity in mice. M2 protein administration shortened the duration of virus shedding and facilitated the recovery of mice after lethal challenge. Okuda et al. (5) administrated a DNA vaccine comprising full M gene (M1 & M2) and showed heterologous protection in immunized mice. They found that specific anti M2 antibodies and strong CTL responses are attributed in protective activity of DNA vaccine. Tompkins et al. (21) evaluated the efficacy of DNA encoding M2 protein as a prime followed by recombinant adenovirus expressing M2 in a prime-boost regimen. Their results indicated that M2 DNA vaccination can induce cross-reactive antibodies, virus specific T-cell responses and protection against lethal H5N1 challenge.
Jimenez et al. (22) immunized mice with a combination of DNA vaccine encoding codon-optimized NP, M1 and M2 genes of influenza. They showed that using plasmid DNA alone could not protect mice from lethal challenge. To improve plasmid-based vaccine application, formulation of DNA vaccine with molecular adjuvant like HSP family has been suggested (12, 23, 24). HSP70 plays an important role in protein folding, assembly of protein complexes and translocation of proteins across cellular compartments. Other important roles of HSP70s, such as serving as carriers for antigenic peptides and promoting the induction and release of cytokines by different immune cells, have been previously mentioned. The adjuvant effect of HSP70 has been reported previously in different studies when it is fused to or coinjected with target gene. The positive stimulatory effect on B cell responses in a DNA vaccine regimen and enhance in vaccine efficacy have been shown when HSP70 gene fused to the target DNA (23, 24).
In an effort to prepare a universal potent DNA vaccine, Jazi and colleagues constructed a chimeric plasmid harboring the extracellular domain of the influenza A M2 protein (M2e) fused to C-terminal domain of mycobacterium tuberculosis HSP70, HSP70359–610, as a carrier and adjuvant and evaluated its immunogenicity in mice model (25). We previously constructed a pcDNA expression vector encoding M2 gene of human influenza virus (A/New Caledonia/20/99) and evaluated the protein expression in different eukaryotic cells (18). In the present study, we developed a chimerical DNA vaccine encoding M2 and L. major HSP70 (amino acids 221-604).B cell epitopes of different fragments of L. major HSP70 were analyzed using B cell epitope prediction software. The results revealed that HSP70 (221-604) has two times more potential B cell epitopes than CT-HSP (491-604). These findings were in accordance with Rafatee's results (17). They found that the entire fragment of HSP70 induced more antibody responses compared to other truncated forms of HSP70, which was not suitable in the case of Leishmania. In contrast, as antibody response specific to M2 protein is more important than cellular response, in this study we fused M2 gene of Influenza virus to the entire L. major HSP70 (221-604).
Naturally, M2 protein has free N-terminus and its C-terminal is attached to the cell membrane (26), therefore HSP 70 gene was fused to the C-terminal of M2 gene to make M2e domain free and convert a low immunogen to a highly immunogenic peptide. The two gene fragments were connected through BamHI site. This restriction enzyme site sequence (GGATCC) incorporates Glycine and Serine into the chimer protein. These two amino acids and Valine (the first amino acid of HSP), provide the minimum linker (GSV) between M2 and HSP in chimer protein. This linker is expected to be soluble and flexible, and should not disrupt the structure of the link fragments (27, 28). In conclusion, our results demonstrated that the M2-HSP70 chimer protein is successfully expressed in eukaryotic cells as evidenced in immunoflurescent and Western blotting assays using specific anti M2 antibody (Figure 4 and 5). In addition, computational studies of the chimer peptide sequence revealed that the immunodominant epitope of M2 protein was not interrupted by HSP. The immunogenicity of this construct with different formulation should be evaluated in future to find an efficient immunization regimen against influenza viruses.