1. Background
The GB virus C is a newly identified human RNA virus which belongs to the Flaviviridae family (1). The GBV-C genome encodes a polyprotein of approximately 3`000 amino acids that contains at least two structural proteins as Envelope protein 1 (E1) and Envelope protein 2 (E2) and also non-structural proteins NS2 to NS5 (2). It is closely related to the hepatitis C Virus (HCV) but is more common than HCV among healthy blood donors (3). Transmission via the blood-borne route is the commonest mode, although vertical and sexual transmission is well documented (4, 5). GBV-C is distributed globally; its prevalence in the general population is tenfold higher in African countries than in non-African countries (4). Patients on maintenance hemodialysis are at increased risk of the novel GBV-C infection (6). In these patients, GBV-C viremia is persistent for many years with uncommon recovery (7).
Hepatocarcinogenicity of GBV-C is an important key question that remains controversial (1, 4, 8). The reports seem conflicting. A few investigations demonstrated their association with hepatitis and cirrhosis of the liver and possible presence in hepatocellular carcinoma. It was also observed in hematological disorders and malignancies (9). Several reports have noted an association between GBV-C and hepatitis-associated aplastic anemia besides other hepatitis causing viruses (10). Nevertheless, Most of the results showed that GBV-C infections are asymptomatic, transient, and self-limiting, with slight or no elevation of alanine aminotransferase (ALT). These findings suggested that GBV-C itself is not an important cause of hepatitis, and co-infection with GBV-C does not alter the clinical course of community acquired hepatitis A, B or C (4). Meanwhile, interestingly, GBV-C co-infection has a beneficial effect on survival and progression to Human Immunodeficiency Virus Infection (AIDS) in patients infected with Human Immunodeficiency Virus (HIV) (9, 11). Nevertheless, an association between GBV-C and HBV and pathogenesis of NHL (non-Hodgkin’s lymphoma) could propose the potential participation of this viral infection in the pathogenesis of NHL (12). Therefore, further investigations are necessary to provide more information.
2. Objectives
The current study aimed to assess the relationship between the prevalence of GBV-C RNA and that of antibody to the putative E2 protein (anti-E2) among the patients receiving hemodialysis referred to the Fifth Azar Medical Centre in Gorgan, Iran, and also to decide reappraisal to screen blood supplies for transfusion-transmitted infection agents such as GBV-C.
3. Patients and Methods
3.1. Patient Samples
This cross sectional study was carried out in a hemodialysis unit in Gorgan, Iran, from June 2012 to July 2012. The study and sampling was approved by the Ethics Approval Committee of Faculty of Medical Sciences, Golestan University of Medical Sciences under No.1005-900428-03، on July 2012 and the informed consent was obtained from the patients. Plasma from all 149 (census method) patients receiving hemodialysis were aliquoted and stored at -20°C and -70°C until use. Demographic and other medical information were obtained from their medical records.
3.2. RNA Extraction
Briefly, RNA was extracted from 200 µL of EDTA (Merck, Germany) anticoagulated plasma by a commercially available kit (High Pure Extraction Kit; Roche Diagnostics GmbH, Mannheim, Germany). Negative (GBV-C RNA negative plasma) and positive (GBV-C RNA positive plasma) controls were included in each run.
3.3. GBV-C Reverse transcription polymerase chain reaction (RT-PCR)
The nucleic acid was used directly for reverse transcription polymerase chain reaction (RT-PCR) for cDNA synthesis by a commercial Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics GmbH, Mannheim, Germany). The cDNA was synthesized from 1.75 μL of extracted RNA at 25°C for 10 minutes, at 50°C for one hour, and at 85°C for five minutes. Each 10 μL RT master mixture contained 1mM dNTP, 60 μM random hexamer primer, 10 U transcriptor reverse transcriptase, 20U RNase inhibitor, 2.5 μM anchored oligo (dT) primer and 2 μL 5X RT buffer.
3.4. Nested- PCR Reaction
polymerase chain reaction (PCR) amplification was performed for the presence of GB virus C RNA with primers HG1, HG1R for the outer primer pairs (262 bases) and HG2, HG2R for the inner primer pairs (188 bases) from highly conserved regions of 5'untranslated region (5' UTR) of GBV-C PNF2161 isolate based on the reference sequences from GeneBank, as described elsewhere (13, 14). The sequences and positions of the primers used are shown in Table 1. All PCR contamination precautions were observed; and negative controls using sera from subjects with no GBV-C markers were obtained from Digestive Disease Research Institute of Shariati Hospital, Tehran University of Medical Sciences, Iran. Polymerase chain reaction procedure was carefully optimized.
The first-round PCR amplification was performed in a 25 µL reaction volume, containing 1 µL cDNA, 10 pmol of each of the universal outer forward and reverse primers (Table 1), 0.2 mM of each deoxynucleotide (dNTP) (Genet Bio (A type), Korea), 1.25 U of Taq DNA polymerase (Genet Bio (A type), Korea), 1.5 mM MgCl2 (Genet Bio (A type), Korea) and 2.5 μL 10X PCR buffer (Genet Bio (A type), Korea). The reactions were performed in a thermal cycler (PeQlab, Erlangen, Germany). The reagents for the second PCR round were the same as the ones used in the first round but the used template amount was 0.5 µL. For the second PCR round, the PCR master mixture contained: Reactions were performed in a Peq Lab thermal cycler (Primus Advanced 96 thermal cycler, USA) programmed as follows: 95°C for five minutes; 30 cycles of 94°C for 50 seconds, 55°C for 40 seconds, and 72°C for 50 seconds; and 72°C for five minutes. Amplification was also done for the second round; 95°C for five minutes; 25 cycles of 94°C for 50 seconds, 70°C for 40 seconds, and 72°C for 50 seconds with a final extension at 72°C for five minutes.
3.5. Detection
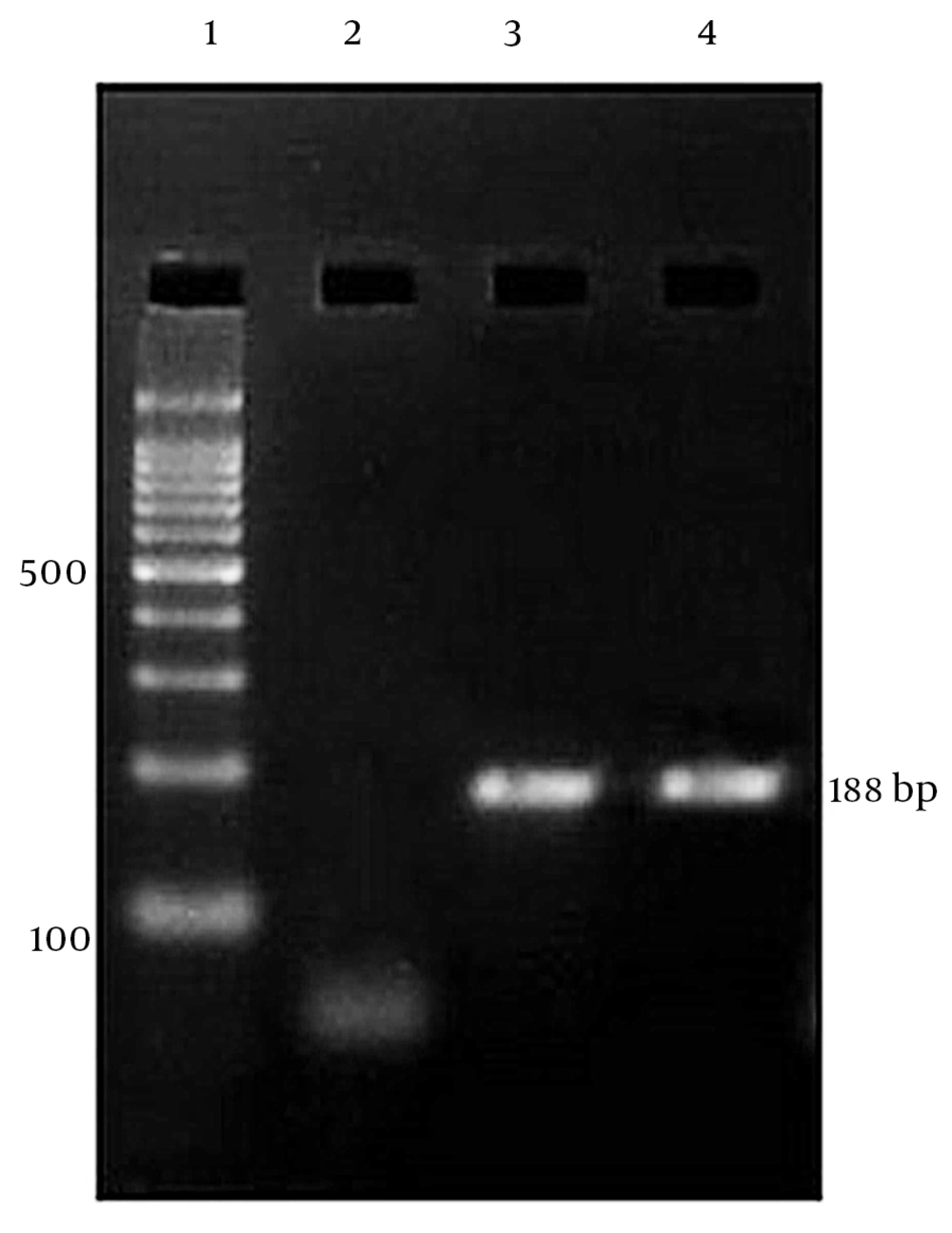
The PCR products were electrophoresed in a 2% agarose gel, stained with ethidium bromide, and visualized by UV illuminator (Figure 1).
3.6. Serological Markers
Anti-E2 antibody of sera was measured by an enzyme linked immunosorbent assay (GBV-C Ab ELISA kit, Rapidtest, USA). Of these, the positive GBV-C RNA + and anti-E2 GBV-C/GBV-C + sera were tested for hepatitis B surface antigen (HBsAg), hepatitis B core antibody (HBcAb IgM), hepatitis C antibody (anti-HCV), and hepatitis E antibody (anti-HEV) by commercial enzyme-linked immunosorbent assay (DS- EIA- HBs-0.01; Fourth Generation Elisa Kit-Italy, DS- EIA-ANTI-HCV; Fourth Generation Elisa Kit- Italy, HBcAb IgM-DiaPlus Inc; Third Generation Elisa Kit- Italy and HEV Ab-DiaPro. Inc., and Third Generation Elisa Kit- Italy respectively). Results were interpreted precisely, according to the manufacturer’s instructions.
3.7. Statistical Analyses
Prevalence and 95% confidence intervals (CI) were calculated. Chi-square test or Fisher’s exact test were performed to evaluate the distribution of characteristics associated with GBV-C infection. Statistical significance was assessed at 0.05 probability level in all analyses. Data were analyzed using SPSS software, version 16.
4. Results
Plasma samples of 149 patients who receive hemodialysis, 74 males, 75 females, mean age: 56 ± 15.92 years, age range: 15 to 90 years, were tested. The total prevalence of GBV-C exposure was 14.7% (95% CI: 0.09-0.21) among the patients receiving hemodialysis, the rates of GBV-C viremia and anti-E2 GBV-C positivity were 6.04% (9/149) and 10.73% (16/149), respectively. Besides, three cases (2.01%) had both markers. In most of the cases (59%), the presence of anti-E2 GBV-C antibody was associated with clearance of GBV-C from plasma. Among 22 subjects who were GBV-C positive, 15 (68.18%) were positive for at least one viral marker, six (27.27%) were positive for anti-HCV, 10 (45.45%) were positive for anti-HBs, 13 (59.9%) were positive for anti- HBc IgM, two (9.09%) were positive for anti-HBc IgM /anti-HCV/HBsAg, and two (9.09%) were positive for anti-HBc IgM /anti-HCV, while the co-infection between GBV-C and HEV was not observed. The levels of liver enzymes were normal in all of them.
There was statistically significant difference between GBV-C exposure and viral hepatitis co-infection and history of addiction (P < 0.00 and P < 0.01, respectively) (Table 2). GBV-C was detectable in 17% of the 51 to 55 year-old cases, and the most cause of renal failure in GBV-C positive patients was diabetic nephropathy (45.45%) (not shown in the table). All 22 GBV-C positive patients had a history of transfusion but one (4/54%) patient. GBV-C exposure did not correlate with gender, age, ethnicity, duration of dialysis, and history of blood transfusions or time on dialysis.
5. Discussions
The new hepatitis G virus/GBV-C was discovered in 1995. The structure of the virus, its genotypes, the clinical features of the disease, routes of viral transmission, and the types of co-infection have been almost completely described (15). The present study represents the first investigation of GBV-C infection among the patients receiving hemodialysis in Golestan Province, North East of Iran. Longitudinal studies on the patients receiving hemodialysis indicated that GBV-C RNA and anti-E2 GBV-C prevalence rates were ranged from 3.1% to 57.5% and 7% to 41.2%, respectively (6, 15-17). These differences in the prevalence of GBV-C infection may also be explained by epidemiological variations, methods of GBV-C RNA detection (especially the use of different primers), duration of dialysis treatment, geographic factors, etc. (18).
Tables 4 and 5 show the frequency of GBV-C viremia, GBV-C seroreactivity, and viral markers observed in the patients receiving hemodialysis, in Iran and the other countries. In the current study the prevalence of GBV-C infection (14.7%) among the patients receiving hemodialysis was found. The rates of GBV-C viremia and anti-E2 GBV-C positivity were 6.04% and 10.73%, respectively. Recent studies in Iran concentrated on some high risk groups and those results focused on detection of one of the viral RNA or serological markers (Table 5). The current study results confirmed a better vision of the infection by detecting both viral RNA and serological markers of GBV-C and the rest of hepatitis virus markers. Therefore, the prevalence of GBV-C contact in patients is underestimated when either antibody or nucleic acid is used. Besides, Tribl et al. identified GBV-C-RNA in PMNC of two patients negative for GBV-C-RNA in serum, suggesting that higher sensitivity assays for determination of the prevalence of GBV-C infection in high risk patients is requisite (19).
In most of the patients, the presence of anti-E2 GBV-C antibody is usually associated with clearance of GBV-C from serum (6). These antibodies seem to provide temporary protection in the patients previously infected with GBV-C (20). In the current study, by examining 149 patients, almost all GBV-C positive samples were either GBV-C RNA positive or anti-E2 GBV-C positive except three (2.01%) who had both RNA and antibody markers, and interestingly , one patient (one out of the three) was not infected with HBV or HCV. This finding does not support Nuebling et al. who believe co-detection of GBV-C viremia and anti-E2 GBV-C antibody occurs only in the patients who have been concurrently infected with other viruses such as HBV, HCV, or HIV (21).
In most of the previous studies, co-infection with HCV in GBV-C- positive patients was observed more frequently than with HBV (Table 3). Yashina et al showed that in patients with acute viral hepatitis B (HBV), and C (HCV) the detection rate of GBV-C RNA was 19%-32% and 20%-48.3%, respectively (22). By contrast, in the current investigation a high frequency of anti-HBs-positive 10 (45.45%) and anti- HBc IgM- positive 13 (59.9%) among the 22 subjects with positivity GBV-C exposure reveals a higher frequency of co-infection with past/ present HBV infection as compared with those of HCV (27.27%). Three out of 22 (13.63%) patients were GBV-C +/anti-HBc IgM + /HBsAg-. During a one year follow-up study, all the three patients showed GBV-C +/ anti-HBc IgM +/HBsAg +. These infections may be associated with low viral loads (23, 24). Thus, the frequent presence of co-infection with HBV in these patients may be underestimated.
In the current study the prevalence of GBV-C exposure was higher in participants aged 51to 55 years (17.2%) compared to the other age groups; this may be a result of increased exposure to GBV-C at older ages (not shown in the table). In several studies, the highest prevalence of GBV-C exposure was reported for individuals aged 29 to 39 years (7). The majority of published findings demonstrated a correlation between GBV-C and history of blood transfusion and duration of hemodialysis (6, 15, 18). In the current study, almost all positivity- GBV-C exposure patients had a history of blood transfusion, but analysis of the results showed no relationship between history of blood transfusion, duration of hemodialysis, and GBV-C positive and negative patients in the current study series. It is suggested that the transmission route for GBV-C may be nosocomial transmission during dialysis; thus, patient-to-patient GBV-C transmission with environmental blood contamination and re-using the dialysis machines may be significant risk factors of GBV-C acquisition in hemodialysis units. Similar results were observed in Germany and England (3, 25, 26).
Even though Food and Drug Administration (FDA) declared GBV-C as a non-harmful virus, this has not been shared by all authors (9). Despite the fact that, no obvious evidence exists regarding the role of GBV-C in liver damage, it appears to play a minor role in acute hepatitis, even in immunosuppressed patients (3, 15). Nevertheless, its effects on kidney injury and graft survival cannot be ignored (27-30). Of course, more studies are needed to confirm these observations.
Investigations indicated that GBV-C RNA has been detected in hepatocytes, peripheral blood lymphocytes and monocytes, vascular endothelial cells, and other tissues (15). Primary replication of GBV-C in organs is questioned and it is suggested that a reservoir of GBV-C could be present in these organs. It seems that this virus could have been re-infected in the absence of protective antibodies (14, 15, 19, 22, 31-35). Therefore, GBV-C infection seems to affect the clinical course of the disease in immunosuppressed patients such as transplant recipients, patients with chronic renal failure and HIV-infected patients that may be responsible for persistent infection or graft rejection. Although no statistically significant relationship was observed between GBV-C exposure and history of blood transfusion in the current study, many investigations indicated a higher risk of infection because of more transfusion of blood and blood products and increasing the chance of infection with blood borne viruses. , Transfusion appears to be an important risk factor for GBV-C transmission in patients; therefore, running a routine screening program for blood donor regarding GBV-C contact seems to be required for immunosuppressed individuals.
In conclusion, regular virological testing and dialysis of GBV-C positive patients in a separate unit can reduce nosocomial GBV-C infection and the prevalence of GBV-C in the patients receiving hemodialysis in the area understudy.
| Primer | Region | Position | Polarity | Sequence (5' to 3') |
|---|---|---|---|---|
| HG1 | 5'UTR | 139 -158 | Sense | 5'-GGTCGTAAATCCCGGTCACC-3' |
| HG1R | 5'UTR | 381 - 400 | Antisense | 5'-CCCACTGGTCCTTGTCAACT-3' |
| HG2 | 5'UTR | 163 -182 | Sense | 5'-TAGCCACTATAGGTGGGTCT-3' |
| HG2R | 5'UTR | 331- 350 | Antisense | 5'-ATTGAAGGGCGACGTGGACC-3' |
| Variable | Total-GBV-C (+) NO = 22 (%100) | Total-GBV-C (-) NO = 127 (% 100) | P-Value |
|---|---|---|---|
| Gender | 0.33 | ||
| male | 13 (59.1) | 61 (48.03) | |
| Female | 9 (40.90) | 66 (51.97) | |
| Ethnicity | 0.24 | ||
| Fars | 15 (68.19) | 102 (80.31) | |
| Turk | 0 | 1 (0.79) | |
| Turkman | 3 (13.63) | 4 (3.14) | |
| Sistani | 4 (18.18) | 18 (14.18) | |
| Cossack | 0 | 2 (1.58) | |
| History of addiction | 0.01 | ||
| Positive | 1 (4.55) | 0 | |
| Negative | 21 (95.45) | 127 (100) | |
| History of blood transfusion | 0.8 | ||
| Positive | 3 (13.64) | 19 (14.96) | |
| Negative | 19 (86.36) | 108 (85.04) | |
| History of hepatitis | 0 | ||
| Positive | 3 (13.64) | 3 (13.64) | |
| Negative | 19(86.36) | 19 (86.36) | |
| Age, y | mean: 54.32 ± 12.56 median: 53 | mean: 55.23 ± 16.47 median: 57 | 0.9 |
| Number | anti-E2 GBV-C | PCR GBV-C | HBs Ag+/GBV-C + | HBc Ab IgM+/GBV-C + | HCV Ab++/GBV-C + |
|---|---|---|---|---|---|
| 1 | + | - | - | + | + |
| 2 | - | + | - | - | - |
| 3 | + | - | - | - | - |
| 4 | + | - | + | + | + |
| 5 | + | - | - | - | - |
| 6 | + | - | - | - | - |
| 7 | - | + | - | + | + |
| 8 | + | + | + | + | + |
| 9 | + | + | + | + | - |
| 10 | + | + | - | - | - |
| 11 | + | - | - | - | + |
| 12 | - | + | - | + | - |
| 13 | - | + | - | - | - |
| 14 | + | - | + | + | - |
| 15 | + | - | + | + | - |
| 16 | - | + | - | - | + |
| 17 | + | - | + | + | - |
| 18 | - | + | - | - | - |
| 19 | + | - | + | + | - |
| 20 | + | - | + | + | - |
| 21 | + | - | + | + | - |
| 22 | + | - | + | + | - |
| Country/City/Year/Reference | Blood Donor GBV-C-RNA+/GBV-C-Anti E2+) | Hemodialysis (GBV-C-RNA+/GBV-C-Anti E2+) | Thalassemic Patients (GBV-C-RNA+/GBV-C-Anti E2+) | HIV Patients (GBV-C-RNA+/GBV-C-Anti E2+) | Heterosexuals (GBV-C-RNA+/GBV-C -Anti E2+) | Intravenous Drug Users (IVDU) (GBV-C-RNA+/Anti E2+) | HCV Patient (GBV-C-RNA+/GBV-C-Anti E2+) |
|---|---|---|---|---|---|---|---|
| Iran/Tehran/2009/(14) | 43.6%/ | ||||||
| Iran/ Tehran/2008/(33) | 10.97%/ | 6.7% | 13.5% | ||||
| Iran/ Tehran/2008/(32) | 1% | ||||||
| Iran/ Tehran/2007/(35) | /3.89% | ||||||
| Iran/ Tehran/2005/(31) | 1%/ | 12.9%/ | 8.8% |
| Country/city/y/Reference | Blood Donor GBV-C -RNA+/ GBV-C -Anti E2+) | Hemodialysis (GBV-C -RNA+/ GBV-C -Anti E2+) | Thalassemic patients (GBV-C -RNA+/ GBV-C -Anti E2+) | HIV Patients (GBV-C -RNA+/ GBV-C -Anti E2+) | Heterosexuals (GBV-C -RNA+/ GBV-C -Anti E2+) | Intravenous Drug users (IVDU) (GBV-C -RNA+/ Anti E2+) | HCV Patient (GBV-C -RNA+/ GBV-C -Anti E2+) |
|---|---|---|---|---|---|---|---|
| Iran/Tehran/2009 (14) | - | - | - | - | - | - | 43.6%/- |
| Iran/ Tehran/2008/(33) | - | - | - | 10.97%/- | 6.7%/- | 13.5%/- | - |
| Iran/ Tehran/2008/(34) | -/1% | - | - | - | - | - | - |
| Iran/ Tehran/2007/(31) | -/3.89% | - | - | - | - | - | |
| Iran/ Tehran/2005/(35) | 1%/- | 12.9%/- | - | - | 8.8%/- | - |