1. Background
Microorganisms such as bacteria and viruses have several components called pathogen-associated molecular patterns (PAMPs). Pattern recognition receptors (PRRs) of host innate immune system such as TLRs can recognize these components during infections (1, 2). Toll-like receptor proteins are members of a big family, which have extracellular domains with leucine-rich repeat (LRR), a transmembrane domain, and a intracytoplasmic region called the Toll/Interleukin-1 receptor (IL-1R) homology (TIR) domain (1, 2). At least 10 members of this family have been identified in mammalian cells, which can recognize specific components conserved among microorganisms (1, 3).
Toll-like receptor proteins are recognized by their ligands. Most of these ligands are found in the pathogens. Therefore, TLR proteins can act as primary stimulators of the innate immune system at the start of microbial infections (1, 3, 4). Pathogen recognition by TLRs stimulates rapid activation of innate immunity by producing proinflammatory cytokines, chemokines, anti-microbial small molecules such as nitric oxide and upregulation of co-stimulatory molecules. Activated innate immunity subsequently leads to the development of effective antigen-specific adaptive immunity (1-3). Therefore, the use of TLR-ligands in vaccines could result in more potent and efficacious vaccines that can activate innate and adaptive immunity (5). Therefore the TLRs are considered to be adjuvant receptors (1, 6).
Flagellin, the major protein constituent of bacteria flagella from both Gram-positive and Gram-negative bacteria, is a mammalian TLR5 agonist. The activation of epithelial cells in response to Salmonella identified flagellin as a stimulatory ligand for TLR5 (7). TLR5 is expressed in epithelial cells, endothelial cells, macrophages, immature DCs, and T cells (2, 8-10).
Both innate and adaptive immunity can be activated by flagellin (11). Numerous studies have demonstrated the effectiveness of flagellin as a systemic and mucosal adjuvant to generate antigen-specific antibodies and to stimulate T cells in mice and nonhuman primates when it is administered either as a native purified protein or as a hybrid protein with physical linkage to target antigens and poorly immunogenic peptides (5, 12-17). Besides, systemic administration of flagellin may be a relatively safe means of providing temporary non-specific protection against a variety of challenges (18).
With this description, DNA encoding the TLR5 agonist (flagellin) as a potent adjuvant, with the ability to effectively stimulate the immune system, can be used either admixed or genetically linked to target antigens, particularly in studies of DNA vaccines. In a recent study, we generated a pVAX-fliC recombinant plasmid consisting of Salmonella entericatyphimurium flagellin, which is a TLR5 ligand and can be used as a genetic adjuvant candidate in eukaryote vector pVAX1 and we evaluated its expression in eukaryotic cells.
2. Objectives
In the present study, the fliC gene of S. enterica subsp. enteric serovar typhimurium ATCC 14028, encoding flagellin protein, was cloned into a the pPrime cloning vector and sub-cloned into a pVAX1 expression vector, and transfected into Hela cells, HEK293 cells and Chinese hamster ovary (CHO) eukaryotic cells. The main goal of this study was to construct the recombinant plasmid pVAX-fliC as an adjuvant candidate for DNA vaccines and evaluate its expression in eukaryotic cells.
3. Material and Methods
3.1. Bacterial Strains, Plasmid, Media, and Culture Conditions
S. enterica subsp. enteric serovar typhimurium (ATCC 14028) was obtained from the American Type Culture Collection (ATCC, Manassas, VA., USA) and cultured on Salmonella Shigella agar medium (Oxoid CM0099, UK ) and Neutrient broth (Himedia, India) at 37ºC and 5% CO2. Escherichia coli strain DH5α (Novagen Inc., Madison, Wis., USA) was used for transformation and the pPrimecloning vector (5PRIME Inc. Germany) was used for cloning the PCR products, while pVAX1 (Invitrogen®, Carlsbad, CA, USA) was used for sub-cloning the fliC gene. Eukaryotic expression vector pVAX1 carries the human cytomegalovirus (CMV) immediate early promoter, the bovine growth hormone (BGH) polyadenylation signal for transcription termination, the kanamycin resistance gene and the pUC origin of replication for maintenance in E. coli. The vector was constructed to be consistent with the Food and Drug Administration (FDA) document.
3.2. Genomic DNA Extraction
Genomic DNA of Salmonella enterica was extracted by High Pure PCR Template Preparation Kit (Roche, Germany), according to the manufacturer’s instruction. Concentration and quality of the genomic DNA were assessed by UV absorbance and electrophoresis on 1% agarose gel.
3.3. PCR Reaction
PCR reaction was performed in a total volume of 50 µl containing 10 ng of DNA, 20 pmol of each primer, 200 µM dNTP, 5 µl of 10X PCR buffer, 1.5 mM MgCl2 and 2.5 U Expand High Fidelity PCR System. A pair of oligonucleotide primers were designed based on fliC DNA sequence that was recorded in GenBank (Accession number NC_016856, Region: 2059063-2060550). Forward primer: 5'-AA GCTAGCGGATCCACCATGGCACAAGTCATTAATACA-3' containing NheI and BamHI restriction sites with a simple Kozak sequence (CCACCATGG) just before the start codon to ensure proper translation of prokaryotic genes in eukaryotic cells and Reverse primer: 5'-AAGCTCGAGGAATTCTTAACGCAGTAAAGAGAGGAC-3' containing XhoI and EcoRI restriction sites with a translation stop codon.
PCR reaction was carried out with 30 cycles of denaturation at 94º C for 1 minute, 58ºC for 1 minute, 72ºC for 2 minutes. The reaction was initiated at 94ºC for 5 minutes as initial denaturation before beginning the PCR cycle, and it was ended with a final extension at 72ºC for 10 minutes in a thermal cycler (TECHNE, UK).Finally, the amplified DNA of fliC gene was visualized by electrophoresis on 1% agarose gel and fliC gene fragments were purified from the PCR product using a High Pure PCR Product Purification Kit (Roche, Germany).
3.4. Gene Cloning
Recovered fliC gene was cloned into pPrimecloning vector by Perfect PCR Cloning Kit (5prime), according to the manufacturer’s instruments. Ligation reaction was prepared in a 10 µl volume containing; 54 ng of fliC gene, 50 ng of pPrimecloning vector, 5 µl of 2X ligation master mix and 2.5 µl of distilled water. This reaction was incubated at 15˚C for 16 hours. The ligation reaction was transformed to E. coli DH5α strain competent cells and dispensed on agar plate containing 50 µg/ mL of kanamycin (Sigma, USA), 200 mg/ mL of Isopropyl β-D-1-thiogalactopyranoside IPTG (Sigma, USA) and 20 mg/ mL of X-Gal (Sigma, USA) to discriminate between recombinant (white) and non-recombinant (blue) cells. The plates were incubated at 37˚C for 16 hours. A number of white colonies were assayed by colony PCR. After selecting the recombinant clones, the recombinant fliC plasmid was extracted from the overnight culture by High Pure Plasmid Isolation Kit (Roche, Germany) according to the manufacturer’s protocol. The recombinant plasmids were detected by restriction digestion with NheI and XhoI enzymes (New England BioLabs, USA) and confirmed by sequence analysis (Bioneer, Korea).
3.5. Subcloning in pVAX1 Eukaryotic Expression Vector
The recombinant plasmid pPrime-fliC and eukaryotic expression vector pVAX1 (Invitrogen, Carlsbad, CA, USA) were digested by XhoI and NheI enzymes (New England BioLabs, USA) at 37˚C for 16 hours. After the enzymes thermal inactivation, the products of digestion were analyzed by electrophoresis on 1% agarose low melting point gel (Roche, Germany) and bands of fliC fragments and digested pVAX1 plasmids were purified from the agarose gel by the Agarose Gel DNA Extraction Kit (Roche, Germany).
The fliC gene was subcloned in the pVAX1 expression vector. Ligation reaction was prepared in a 20 µl volume containing; 300 ng of fliC gene, 200 ng of digested pVAX1, 2 µl of 10X ligation buffer and 1 µl of T4 DNA ligase enzymes (New England BioLabs, USA). This reaction was incubated at 15˚C for 16 hours. The recombinant pVAX-fliC plasmid was transformed into competent E. coli DH5α cells and recovered in super optimal broth with catabolite repression (SOC) free of antibiotic at 37˚C for one hour. Next, the cells were cultured on LB agar medium containing 50 µg/mL of kanamycin (Sigma, USA), and incubated at 37˚C for 16 hours. A number of colonies were assayed by colony PCR and recombinant plasmids with the correct restriction pattern were selected by restriction digestion with NheI and XhoI enzymes (New England BioLabs, USA) and used for DNA sequencing with the T7 Forward and bovine growth hormone (BGH)reverse primers (Bioneer, Korea) to confirm that fliC gene is cloned in the proper orientation.
3.6. Expression of Recombinant Flagellin in Mammalian Cells
Hela cells and HEK293 cells were cultured in Dulbecco's Modified Eagle's medium (DMEM) (GIBCO-BRL) supplemented with 10% fetal calf serum (FCS) in the presence of penicillin G (100 U/ mL), streptomycin (100 µg/ mL), gentamycin (50 µg/ mL) and amphotericin B (2.5 µg/ mL) (GIBCO-BRL) at 37˚C with 5% CO2 and transiently transfected with either pVAX-fliC or pVAX1 by electroporation. Briefly, growing cells were trypsinized, spun down, and resuspended in HeBS buffer (25 mM Hepes pH 7.1, 140 mMNaCl, 5 mM KC1, 0.75 mM Na2HPO4) at cell concentration of 5×106/ mL containing 100 µg/ mL plasmid, and electroporated at room temperature in a 2 mm gap cuvette with 200 µL of sample, exponential decay waveform of 160V pulse and 500 µF capacitance by Gene PulserXcell electroporation system (BIORAD-USA).
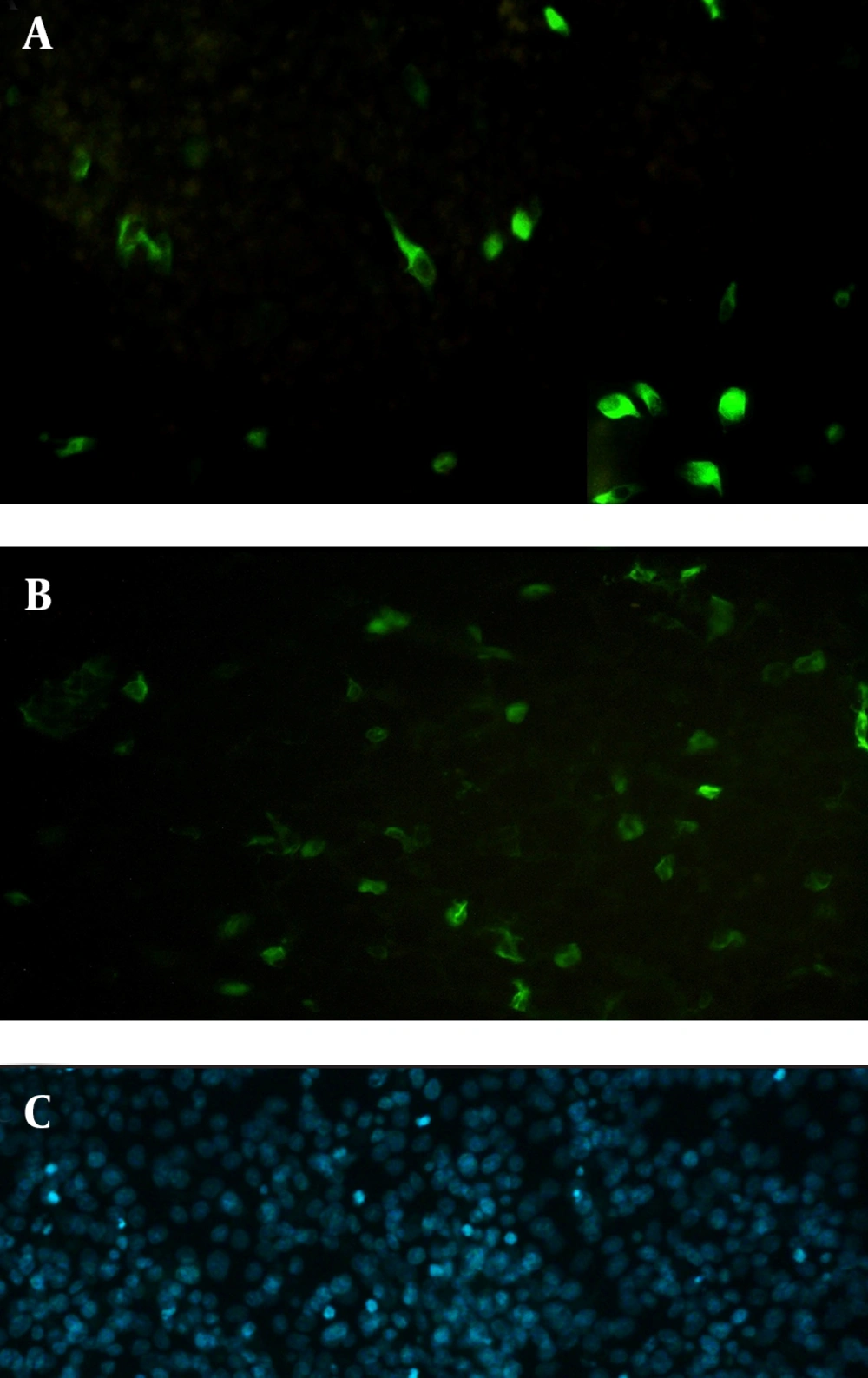
Electroporated cells were cultured in six-well culture plates (Nunc, Denmark) at 37˚C in a 5% CO2 environment. Forty-eight hours after transfection, the cells were fixed with cold methanol–acetone (1:1) for 20 minutes, then rinsed and blocked with 2 mL of 0.15 M phosphate buffered saline (PBS) containing 1% bovine serum albumin (BSA). Flagellin expression was detected using the indirect immunofluorescence assay (IFA), with 2.5 µg/mL mouse anti-fliC (flagellin) antibody (Biolegend, USA) followed by staining with 10 µg/mL fluorescein isothiocyanate (FITC)-labeled anti-mouse IgG (Abcam, USA). Next, 1 µg/mL 4',6-diamidine-2'-phenylindole dihydrochloride (DAPI) (Roche, Germany) was used for staining the nuclei of the cells. After rinsing with PBS, cover slips were immediately observed under a fluorescence microscope (Nikon, USA).
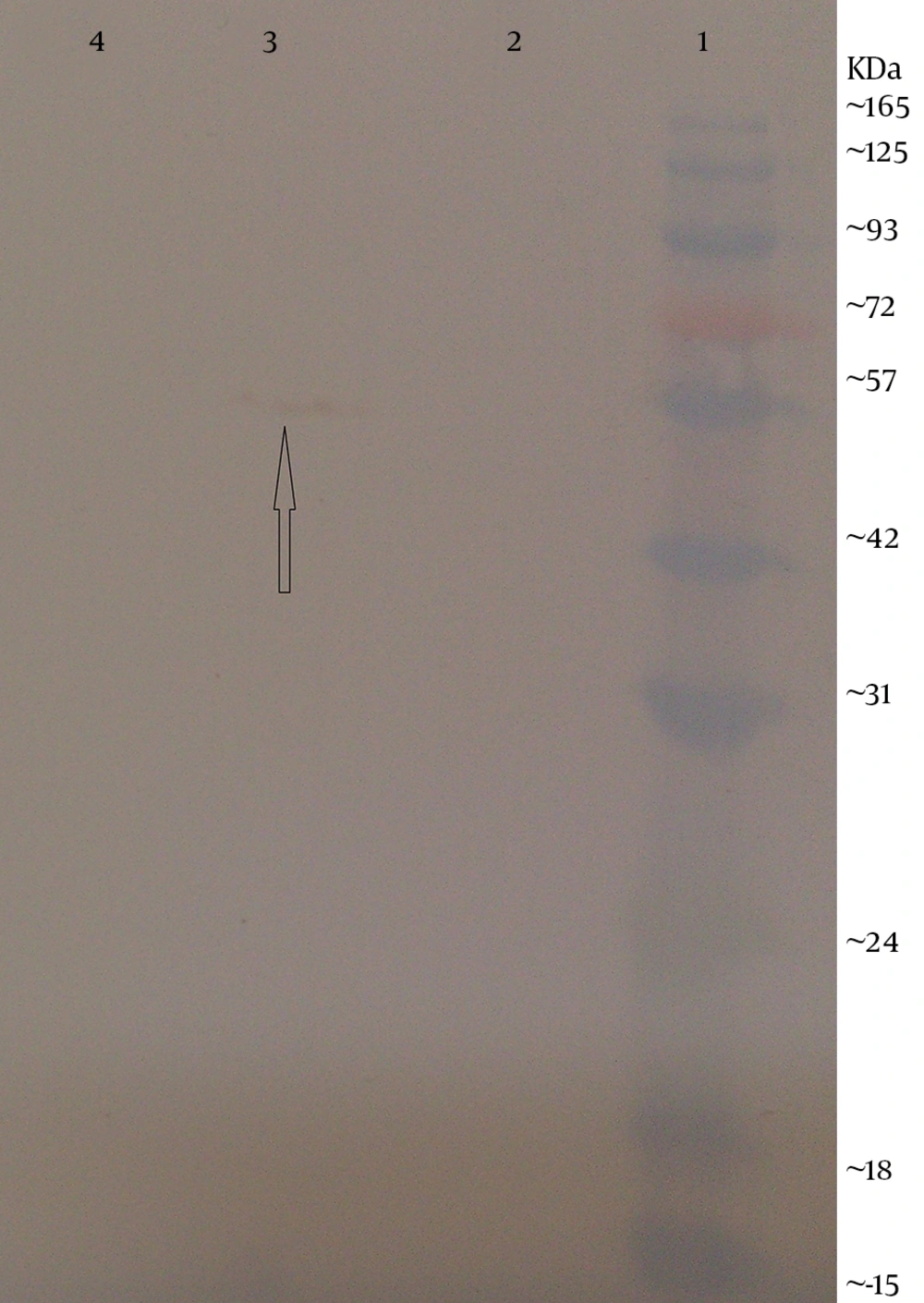
The expression of flagellin in electroporated CHO cells and HEK293 cells was identified by western blotting. The transfected cells were maintained in DMEM-F12 (GIBCO-BRL) at 37˚C and 5% CO2 atmosphere. After 48 hours of incubation, the cells were collected and boiled in the sodium dodecyl sulfate (SDS) sample buffer (60 mMTris–HCl pH 6.8, 2% SDS, 1 mM Ethylene diaminetetraacetic acid (EDTA), 10% glycerol, 0.005% bromophenol blue and 5% β-mercaptoethanol) for 10 minutes. Proteins of transfected cells were separated using 10% (w/v) SDS-PAGE gel. Following electrophoresis, the resolved proteins were blotted onto a 0.45µm pore polyvinylidene fluoride (PVDF) membrane (Roche, Germany) in towbin buffer (0.025 M tris, 0.192 M glycine (pH = 8.3) with 20% methanol) using a semi-dry transfer cell (Bio-Rad, USA) at 15V for 1 hour and the membrane was blocked for 1 hour at room temperature with 3% (w/v) BSA diluted in tris-buffered saline (TBS) (50 Mm tris pH = 7.5, 150 mM NaCl). The membrane was incubated with 1 µg/mL mouse anti-fliC (flagellin) antibody (Biolegend, USA) overnight at 4◦C. After washing with TBS containing 0.1% tween 20, the membrane was stained with diluted goat anti-mouse IgG horseradish peroxidase (HRP)-labeled secondary antibody (1:2,000; Biolegend, USA) for 1 hour. The reaction was detected using 3, 3’-Diaminobenzidine (DAB) reagents (Sigma-Alderich, USA).
Transcription of flagellin gene at the level of mRNA in CHO and Hela cells was also identified by RT-PCR. Total RNA isolation from the cells by RNeasy Mini Kit (Qiagen, Germany) and RT-PCR was performed according to the manufacturer’s protocol using a QIAGEN One Step RT-PCR Kit (Qiagen, Germany) by specific primers (forward primer: 5'-CGTATCAACAGCGCGAAAGA-3' and reverse primer: 5'-CTGGATGGAGTCGAGGTCAG-3'); the final product was then electrophoresed on 2% agarose gel.
4. Results
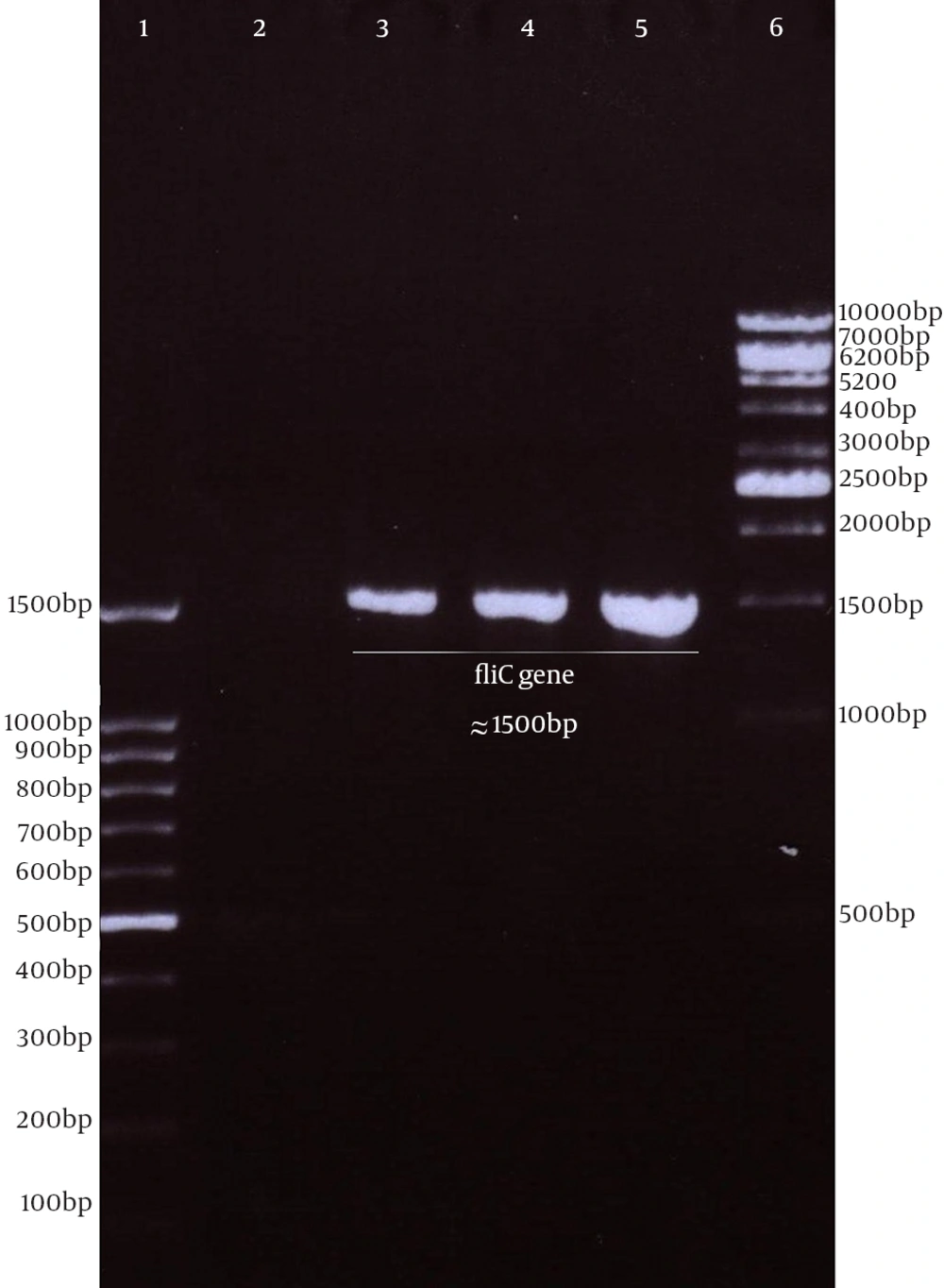
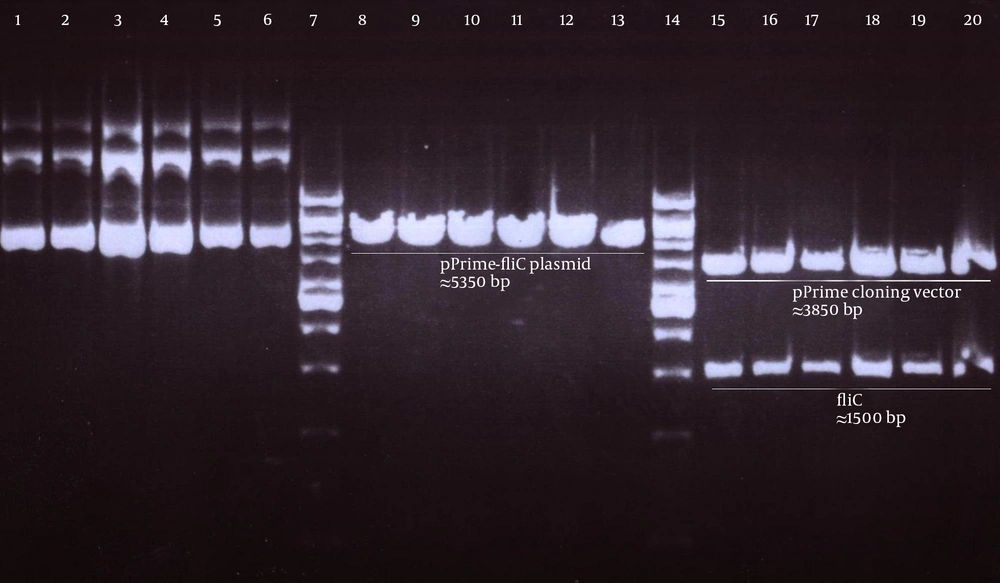
Genomic DNA of S. enterica serovar typhimurium extracted using a commercial kit showed a concentration of 170 µg/ mL and good quality on agarose gel. The full protein-coding region of the fliC gene of S. typhimurium was amplified using designed specific primers. The size of PCR product on agarose gel electrophoresis for fliC gene was 1.5 kbp (Figure 1). The PCR products were cloned into a pPrime cloning vector and this resulted in a recombinant pPrime-fliC plasmid that was transformed successfully into E. coli bacteria. The recombinant plasmids from the white colonies were PCR positive against the fliC gene and vector primers. The recombinant pPrime-fliC plasmids were confirmed by double digestion with NheI and XhoI restriction enzymes (Figure 2). Sequencing analysis of the constructed plasmid also confirmed no amplification errors in sequence of the cloned fliC gene.
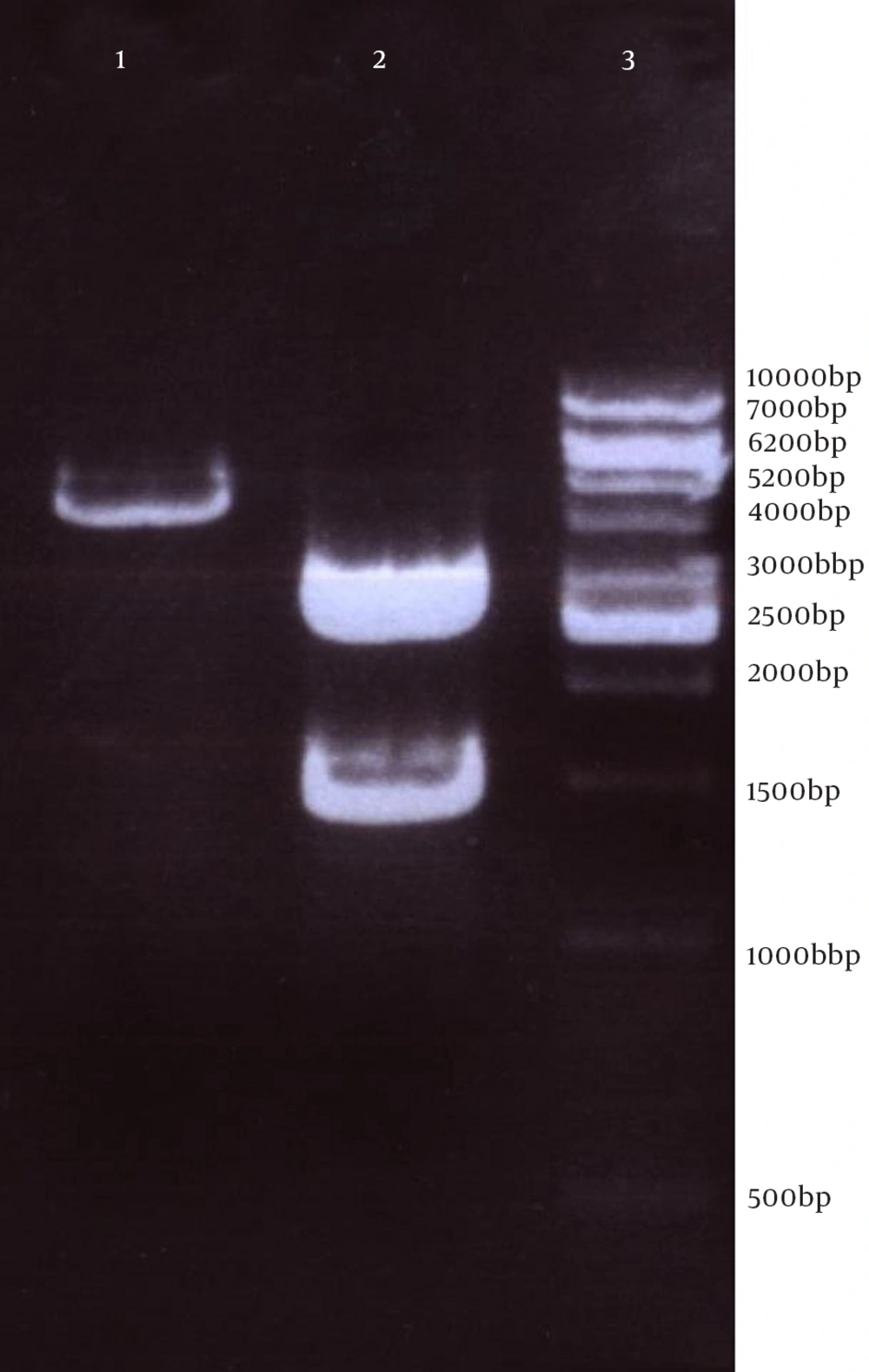
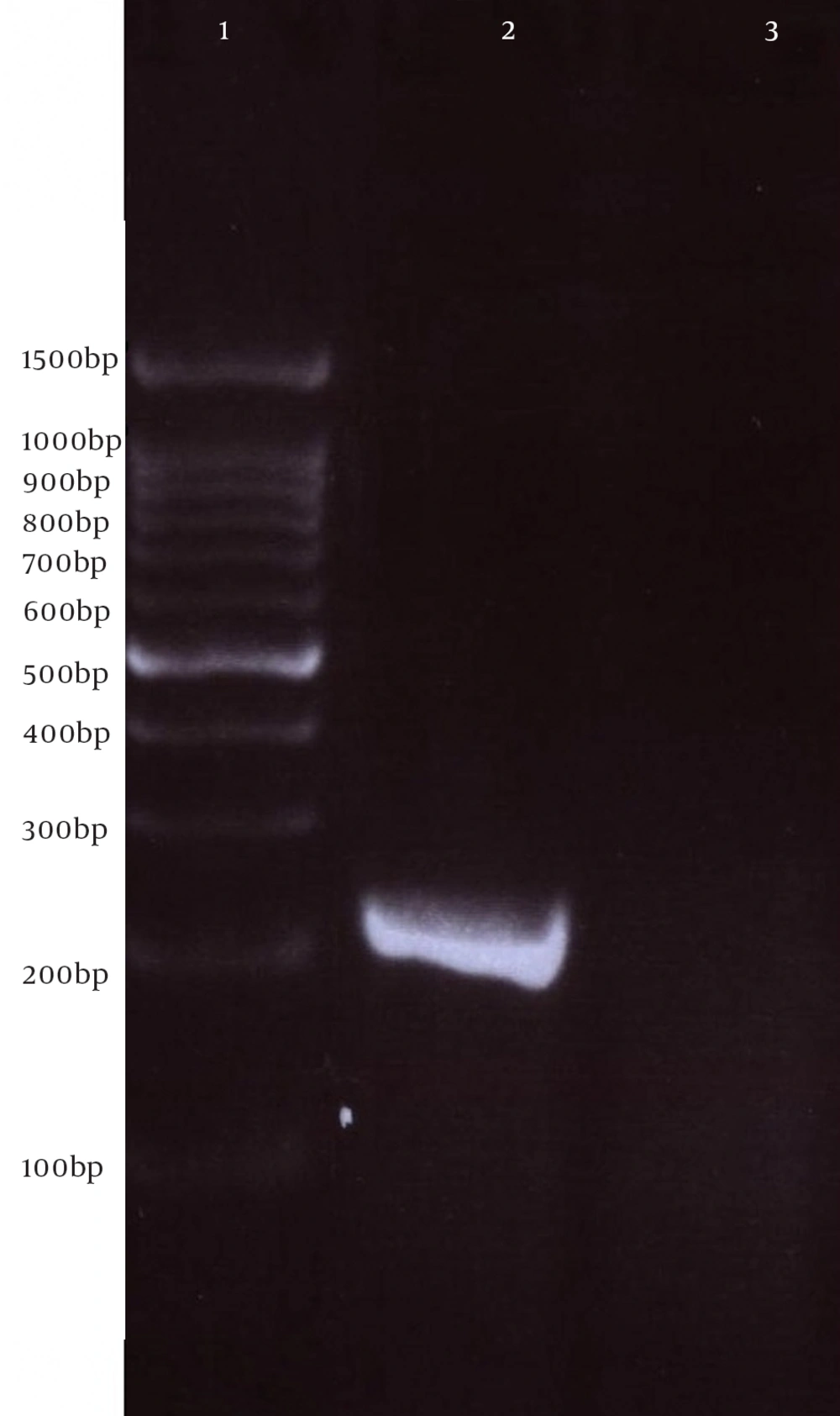
Next, this pPrime-fliC plasmid was digested by NheI and XhoI restriction enzymes and fliC gene was successfully inserted between the NheI and XhoI restriction sites of the pVAX1 expressing eukaryote vector and the pVAX-fliC plasmids were transformed into E. coli DH5α cells. The recombinant pVAX-fliC plasmids were detected by digestion with NheI and XhoI restriction enzymes (Figure 3) and were then confirmed by sequence analysis and compared with fliC sequences in the GenBank database. The fliC gene sequence was 100% identical to S. enterica subsp. enteric serovar typhimurium ATCC 14028, accession no.: NC_016856. In order to examine the in vitro expression of the flagellin protein, eukaryotic expression vector pVAX-fliC was electroporated successfully into Hela cells, HEK293 cells and CHO cells. Transcription of fliC mRNAs was confirmed by RT-PCR method; finally the results were electrophoresed on 2% agarose gel, which identified a band 231 bp (Figure 4). Expression of flagellin protein in Hela cells and HEK293 cells was evaluated by IFA, 48hours post-electroporation (Figure 5). Also, expression of the flagellin protein was evaluated by western blotting with cell lysates of transient transfected CHO cells and HEK293 cells. The molecular weight of the flagellin protein was about 52 kDa (Figure 6).
5. Discussion
Production of a strong immune response and induction of both humoral and cellular immunity against vaccine antigens strongly depend on a formulation with adjuvants. Immunologic adjuvants are agents incorporated into vaccine formulations to enhance the immunogenicity of vaccine antigens (19). Many natural and synthetic compounds as immunological adjuvants including inorganic salts such as aluminum hydroxide, nano and micro particles, oil emulsions, cytokines and microbial compounds, have been studied in vaccine researches (19, 20). The use of microbial-adjuvants in human prophylactic vaccines which can induce the production of endogenous cytokines have priority over the use of recombinant cytokines as adjuvants that have high incidence of local and systemic adverse side effects (21).
A primary mechanism of action for most microbial-based immunological adjuvants is to present pathogen-specific molecular patterns (PAMPs) in association with protective antigens triggering pathogen recognition mechanisms and the activation of innate immunity. The recognition of PAMPs in mammalian cells is mediated by innate immune receptors such as pattern recognition receptors (PRRs), which are expressed by antigen-presenting cells (APC). However, activation of the PRRs such as TLRs not only induces strong inflammatory responses but also stimulates protective adaptive immune responses (3). Recent advances in the field of innate immune system have disclosed cellular and molecular mechanisms behind the adjuvant effects of PAMPs and have led to the use of TLR ligands as adjuvants (1, 6). TLRs are a type of PRR which react with PAMP-based adjuvants or microbial adjuvants. Therefore, the use of TLR agonists such as flagellin (a bioactive TLR5 ligand) as adjuvants is a promising approach to enhance the effectiveness of vaccines and may be the most useful for vaccines that may lack sufficient recognition by host innate immune mechanisms.
Flagellin, a TLR5 ligand, is the major protein component of bacterial flagella, which provide motility and chemotaxis, and allow adhesion to host mucosal tissues (22). The TLR5 ligand is a potent T-cell antigen and has potential as a vaccine adjuvant. Unlike other TLR agonists, flagellin tends to produce mixed Th1 and Th2 responses rather than strong Th1 responses. Additionally, in contrast to other vaccine adjuvants, such as complete Freund's adjuvant, flagellin may exert strong adjuvant effects on both humoral and cellular immune responses following administration through mucosal routes (12). Flagellin can be used as an adjuvant mixed with the antigen but it is more frequently fused to a recombinant vaccine antigen (5, 11, 12). In addition to the upregulation of TLR5 expression, TLR3 expression was also found to be activated by flagellin, thus it could be auseful against for RNA viruses (16). Flagellin is a strong activator of a broad range of cell types involved in innate and adaptive immunity. Moreover, Delaney et al. established that flagellin can promote robust responses against weakly immunogenic antigens of vaccinia virus (23). The efficacy of preventive vaccines against infectious diseases declines dramatically with the progression of age among older adults (24). Adjuvants which lead to an increase in immunogenicity, are required to stimulate the appropriate immunity in the elderly. Bates et al. indicated that flagellin can promote adaptive immune responses in aged mice (13).
According to the study of Vijay-Kumar et al., systemic treatment of purified flagellin in mice did not induce serological, histopathological and clinical hallmarks of inflammation that are induced by lipopolysaccharides, yet protected mice against chemicals, pathogens and ionizing radiation (18). In addition, bacterial genes typically contain immunostimulatory unmethylated CpG motifs (PuPuCpGPyPy) within their DNA backbone (25). We Investigated the CpG motifs in the entire sequence of the fliC gene and observed that the fliC gene’s sequence contains four immunostimulatory CpG motif sites, including agcgtc at positions 89-94, aacgct at positions 199-204, agcgcc at positions 353-358 and aacgtc at positions 1465-1470 (based on GenBank accession number NC_016856, Region: 2059063-2060550).
The aim of the present study was to construct the expression plasmid pVAX-fliC and evaluate its expression in eukaryotic cells. Initially, the full-length of fliC gene from S. typhimurium was amplified using specific primers containing the Kozak sequence and cloned in thepPrime vector and then sub-cloned in expression plasmid pVAX1. Next, cloning and subcloning was confirmed by PCR, restriction enzyme digestion and DNA sequencing. The DNA sequencing results also confirmed the authenticity of the cloned flagellin gene sequences. Following this step, mRNA synthesis of flagellin in eukaryotic cells transfected with pVAX-fliC was confirmed by the RT-PCR test. Also, the presence of flagellin protein was determined in these cells by immunofluorescence assay and western blotting using a monoclonal antibody. Better results were seen for the immunofluorescence assay with HEK293 cells compared to Hela cells. Therefore, it is better to use HEK293 cells to evaluate the expression plasmid. Western blot analysis showed a protein band slightly lower than the 57 kDa band of the protein marker, which was in agreement with the expected molecular weight of flagellin protein (≈52 kDa). The results of this study indicated that the flagellin gene was successfully cloned into the expression plasmid pVAX1, and the expression of this gene at the level of transcription and translation was confirmed in eukaryotic cells. The construction of the pVAX-fliC recombinant plasmid is the first step towards the use of this plasmid as a genetic adjuvant candidate in vaccine studies in the future.
Applequist et al. cloned the membrane-associated form of flagellin in expression vector pcDNA3.1 (26). However, in our study, the full-length fliC gene was cloned in the pVAX1 expression vector, and the sequence of the transmembrane domain or secretory signal sequence was not used. This is because the immune response to antigens that are on the cell surface may be different from the antigens, which are not on the cell surface and could play a role in the polarizing immune responses and raising different profiles of T-cells (27). The adjuvant effect of flagellin has been shown by a variety of pathogen models. An increasing number of studies have demonstrated that flagellin contributes to the efficacy of various novel vaccines for Yersinia pestis (15), West Nile virus (28), Plasmodium falciparum (29), Schistosoma mansoni (30), Tetanus toxoid (31) and influenza (17, 32-34). Flagellin activates TLR5-positive dendritic cells (DCs), neutrophils and epithelial cells (2, 35). Therefore, a powerful vaccine can be created by fusion of flagellin with an antigen in order to enhance the immunopotency of the antigens (19). However, in most mentioned studies they used flagellin as a recombinant protein for vaccine adjuvants, however in this study, for the first time, we inserted fliC gene in pVAX1 for DNA vaccine as an adjuvant candidate.
In conclusion, the recombinant plasmid of pVAX-fliC that was successfully constructed in the present study could be used either for cocktail or physical linkage to target antigens as an effective DNA microbial-adjuvant for future DNA vaccine studies against infectious diseases.