1. Background
Bacterial infection in burned patients is a major problem for health care settings because of the increasing level of mortality and hospitals cost (1). Gram-negative bacteria including Pseudomonas aeruginosa, Acinetobacter baumanii andKlebseilla pneuomoniae are the most frequent pathogens isolated from burned infected wounds (2-4). In 2008, the antibacterial resistance pattern of aerobic bacteria isolated from burn patients showed that ceftazidime and cefotaxime resistant Pseudomona spp. is a common etiological agent of infections (5). In 2010, A. baumanii isolated from burned patients contained carbapenemase genes in their genome (6). Generally, many mechanisms are responsible for antibiotic resistance in Gram-negative bacteria. Relatively, mobile gene elements such as transposon, integrons and plasmids, play important roles in antibiotic resistance character (7-9).
2. Objectives
The present study was conducted to evaluate the antibiotic resistance patterns and its relation with the existence of plasmid in Gram-negative bacteria isolated from burned patients in Isfahan burned hospital, Iran.
3. Materials and Methods
3.1. Bacterial Strains
A total of 139 non-duplicate bacterial isolates were collected from two hundred twenty patients from March to June 2012. Samples were taken at the beginning of the first bondage during the hospital stay (10). The phenotypic features of isolates were identified by standard biochemical tests such as Gram staining, sugar fermentation and API 20E system (Biomerix Co, France).
3.2. Determination of Antibiotic Susceptibility of the Isolates by Disc Diffusion Method
Determination of antibiotic susceptibility patterns were carried out by the disc diffusion method according to CLSI guidelines (11). To perform the test, antagonistic bacteria viz., Escherichia coli ATCC 25922, P. aeruginosa ATCC 27853, Staphylococcus aureus ATCC 25923 and Enterococcus faecalis ATCC 29212 cultured in nutrient broth and their turbidity were adjusted to 0.5 McFarland standard. The suspensions were streaked onto Mueller-Hinton agar plates and the following antibiotic disks (Hi-Media, India) amikacin (30 µg), cefotaxim (30 µg), ciprofloxacin (5 µg), imipenem (10 µg), meropenem (5 µg), co-trimoxazole (25 µg), piperacillin/tazobactam (25 µg), ceftriaxone (30 µg), amoxicillin-clavulanic acid (30 µg), ceftazidime (30 µg) were placed on the seeded media. The plates were incubated at 37°C for 18-24 hours and then the inhibition zone around each antibiotic disk was measured and recorded.
3.3. Plasmid Isolation
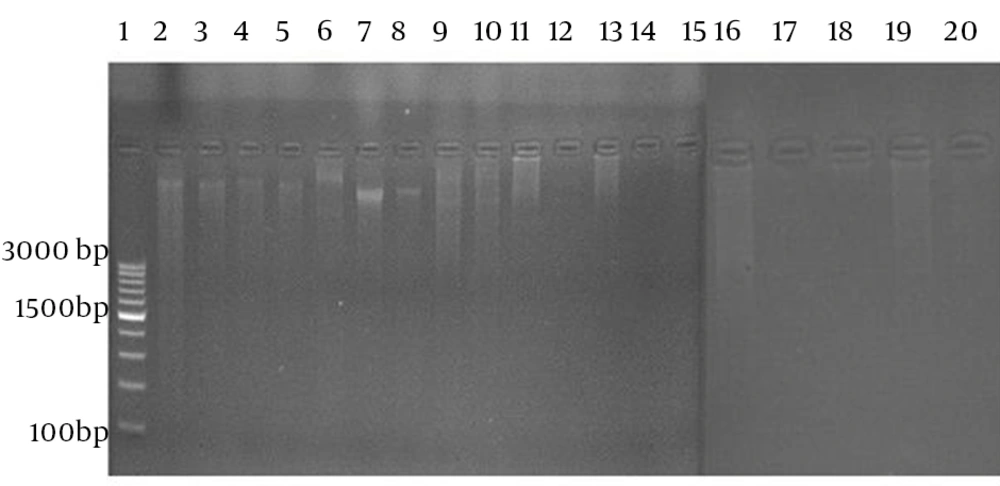
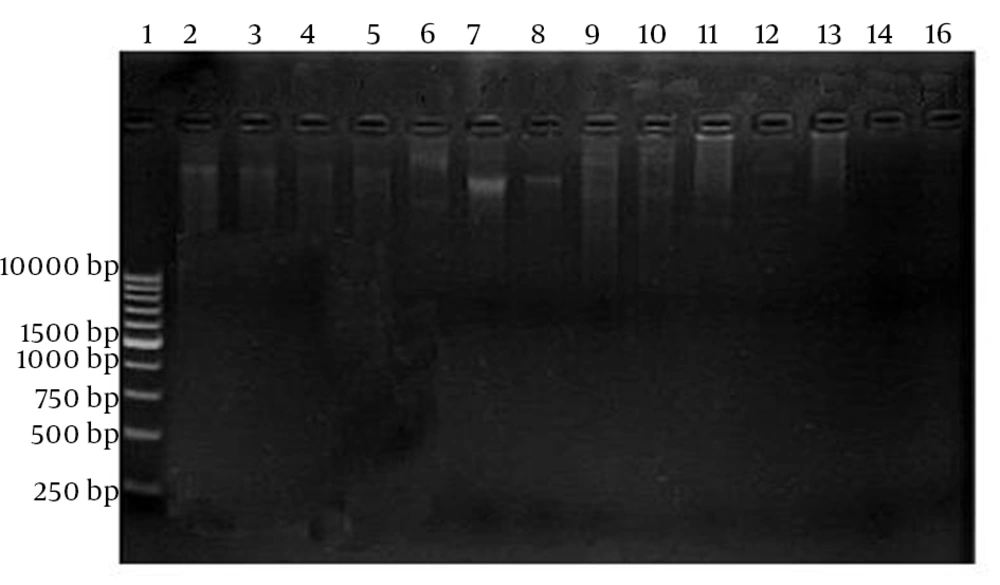
Plasmid was isolated using standard method (Vivantis nucleic acid extraction kit, GF-1 model, version 2.2, Germany) recommended by Birnboim et al. (12) and the purity of plasmid was evaluated at 260 nm by spectrophotometer (Eppendorf Biophotometer 6131, Germany). The plasmid DNA was observed by gel electrophoresis. To prepare the gel, Agarose gel powder (2%) was dissolved in TAE buffer (40 Mm Tris-Hcl, 50 mM Sodium acetate, 1 Mm EDTA; pH 8). The extracted DNA was added to the gel and the Gel was run for two and half hours at 50 V, stained for 30 min with ethidium bromide (0.5 µg/mL). The plasmids were visualized under UV light in Alpha imager gel documentation system (Syngene, UK) (Table 1).
3.4. Plasmid Curing
The isolates were showing resistance character (100%) subjected to plasmid curing. Ethidium bromide was serially diluted in Muller Hinton broth. The curing agent was tested at 500, 250, 125, 62.5, 31.2 µg/mL concentrations. Overnight growth culture of five isolates subjected to plasmid curing and each of them inoculated into the tube containing 1 mL Muller Hinton broth and the incubated at 44°C for 24 hours. The minimal inhibitory concentration (MIC) of Ethidium bromide was determined then the highest concentration permitting growth (SIC) was considered as plasmid curing. The overnight culture was inoculated on Mac Conkey agar with sic of curing agents and incubated at 37°C for two days. The single colonies were picked up by sterile toothpicks and inoculated on nutrient agar plate (30 colonies/plate).The plates was incubated at 37°C for 24 hours and used as master plates. These colonies were inoculated on nutrient agar with ciprofloxacin (5 µg) and on nutrient agar with meropenem (5 µg) and on nutrient agar with ceftriaxone (30 µg) and then incubated at 37°C for 48 hours. The colonies that did not grow on selective medium, were known as cured colonies. At the same time, a non-cured culture on selective medium as a control for curing of each marker was performed. The loss of plasmid DNA was confirmed by gel electrophoresis.
4. Results
4.1. Identification and Antibiotic Susceptibility of the Isolates
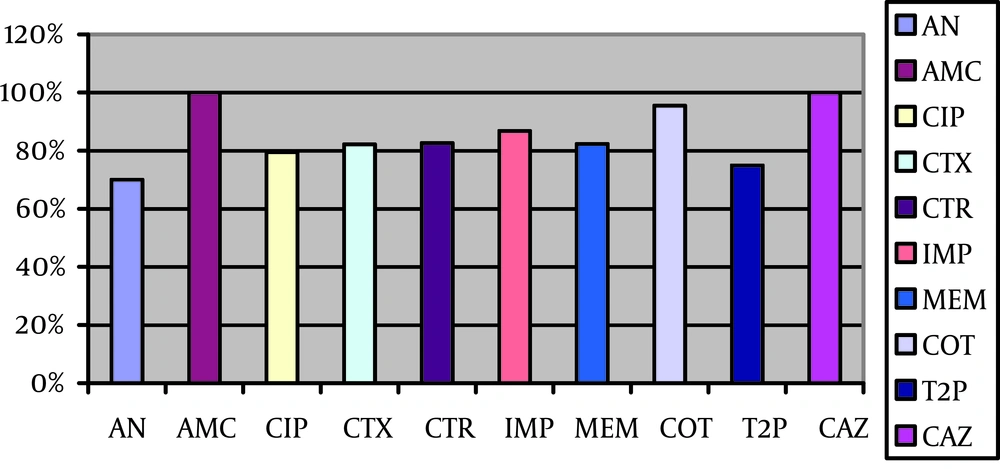
Totally 117 Gram-negative bacteria were isolated and the most common were P. aerugionsa (37.6%), P. fluorescens (25.6%), A. baumanii (20.5%) and K. pneumoniae (7.6%), respectively. The isolates were resistant against ceftazidime and co-amoxiclave with high frequency (100%) and against amikacin with low frequency (70%) (Figure 1).
4.2. Plasmid Isolation
Nineteen resistant strains against all antibiotics were subjected for plasmid isolation (Figure 2).
4.3. Plasmid Curing
Five isolates of bacteria that showing resistance to all antibiotics were selected for plasmid curing. The results showed that A. baumanii and P. aeruginosa were cured (with 60% frequency) whereas; P. fluorescens was not cured (60%).
| Bacteria of Isolates | SIC, µg/mL | MIC, µg/mL |
|---|---|---|
| A. baumanii | 62.5 | 125 |
| A. baumanii | 250 | 500 |
| P. flourescens | 1000 | 2000 |
| P. aeruginosa | 1000 | 2000 |
| A. baumanii | 125 | 250 |
4.4. Determination of Plasmid Mediated Antibiotic Resistance
Our findings showed that 83% to 66% of the cured strains were sensitive to ciprofloxacin and ceftriaxone. However, 33% of the isolates were sensitive to meropenem (Table 2 and Figure 3).
| Antibiotics | Susceptibility of Cured Strains, % |
|---|---|
| Ceftriaxone | 66 |
| Meropenem | 33 |
| Ciprofloxacin | 83 |
5. Discussion
Recently, high frequency antibiotic resistance bacteria, especially in the hospitals, culminated a serious problem for treatment of infectious diseases. In this regards, several resistant strains of S. aureus, K. pneumonia and P. aeruginosa strains against ceftazidime and imipenem and carbapenem respectively were isolated from hospitals (3, 13). Parallel to these reports the present study isolated antibiotic resistant Gram-negative bacteria from burned patient in hospitals. Some of the isolates showed resistance to ceftazidime and Amoxicillin-clavunic acid. In a similar study P. seudomonas aeruginosa isolated from burned patient in Sanandaj, Iran were 100% resistant to ceftazidime (14). On the other hand, parallel to our finding several reports showed that amikacin has effective antibiotic featuers against burned infectious agents (15-17).
In the present study, the most common Gram-negative bacteria isolated from the patients were P. aeruginosa, Acinetobacter spp and K. pneumoniae. Out of all, Acinetobacter spp and P. aeruginosa isolates exhibited common antibiotic resistance pattern (3, 18, 19). In different investigation in Brasilia P. aeruginosa, A. baumanii and K. pneumoniae were introduced as collective nosocomial infectious agents (20). Generally, antibiotic resistance markers located in the chromosomes, plasmids, integron and transposons (21). Of all, antibiotic resistance markers encoded in plasmid play an important role for transferring the resistance marker among different bacteria (22, 23). In the present study 60% of the strains harbored plasmid and they showed resistance property to ciprofloxacin and ceftriaxone.
In 2003, 95% of antibiotic resistant P. aeruginosa isolated from burn patients in Tehran hospitals harbored plasmid (24). In addition, plasmid curing confirmed the relationship between antibiotics resistance character and the existence of plasmid (25). In the present study cured A. baumanii and P. aeruginosa lost their resistance markers after losing their plasmids. Therefore, it can be interpreted that probably antibiotic resistance markers of these bacteria are plasmid mediated. Overall, our investigation showed the existence of the resistance markers in the plasmid of isolated bacteria from burned patients. Hence these markers might easily transfer among the bacteria in the hospitals.