1. Background
Hepatitis delta virus (HDV) was first identified by Rizzetto (1). The presence of Pro-205 in HDV genotype 1 may account for its higher assembly efficiencies and wider distribution (2). New studies suggest at least eight HDV clades, which are phylogenetically distinct and have predominance in different regions worldwide. Clade 1 of HDV is found in all regions. However, Clade 2 is mostly reported from Japan, Taiwan, and Russia. Clade 4 is localized to Taiwan and Japan. Clade 3 has the highest prevalence in the Amazonian region while HDV clades 5, 6, 7 and 8 are found in Africa (3). The epidemiology of HDV infection has been classified into three categories as follows: 1. the endemic pattern that exists in countries such as Greece and Italy; 2. the epidemic pattern that is observed in Venezuela; and 3. the occurrence of HDV infection that is found among intravenous drug users in developed countries (4).
The endemic pattern of HDV infection exists in the Middle East (4). Globally, HDV infection is found in more than 15 million people. Its prevalence is mostly focused in Italy, Eastern Europe, and western Asia (5). Recently, a study reported that there are about 350 million carriers of HBV infection worldwide of which 18 million are infected with HDV (6). Countries such as Iran and Pakistan showed an increase in HDV prevalence. On the other hand, countries like Turkey, India, Australia, China, Japan, and Taiwan, which had a very high HDV prevalence in the past, have shown a decline in the incidence rate, although a high prevalence rate persists in some of them (6). Alavian et al. reported the prevalence of HDV infection in different provinces of Iran. They reported the prevalence rate of 9.7% in Fars, 2% in Yazd; 11.5% in HBsAg positive subjects and 63.7% among cirrhotic patient in Khuzestan; 20.7% in Kerman; 13.8% in Sistan and Balochestan; 5.8% in Golestan; 2.4–5% in Tehran; 9.3% in East Azarbaijan; and 31.37% in patients positive for HBV and HDV infections in Kermanshah (8, 9).
In Iran, the prevalence of HDV is considered to have declined from 18.03% in 1990 to 5.7% in 2005. However, HbsAg positivity is a serious risk for HDV infection (7, 9, 10). Malekzade et al. reported the first case of HDV infection in Iran (11). It occurred as a co-infection or superinfection with hepatitis B virus (HBV). The persistent replication of HBV and HDV was associated with more severe outcomes. Over 90% of patients who are superinfected are likely to develop severe conditions such as cirrhosis or hepatocellular carcinoma. Coinfection with HBV is responsible for more cases of acute liver disease and fulminant hepatitis (70–80% of the total cases) (12-14), while it evolved to chronicity in only 2% of cases (14).
Longitudinal follow-up studies have indicated that the HDV genotype 1 is the most critical factor associated with survival rate along with severity and progression of liver failure (1, 15). Genotype 2 is associated with a milder hepatitis and 2a is more pathogenic (16). Genotype 3 often leads to fulminant hepatitis (16). The frequency of HDV genotypes and clades were undetermined in the city of Mashhad. In consideration of the geographical vastness of Mashhad, which is home to many immigrants from Afghanistan as well as being a city of pilgrimage attracts many people from different regions of Iran and other middle-eastern countries. There, it is expected that the pattern of prevalence is different from other parts of the country.
2. Objectives
This study was performed to determine the frequency of HDV genotypes in the city of Mashhad, Iran.
3. Patients and Methods
Twenty-five subjects were enrolled in this study. Informed consent was obtained and a questionnaire was filled for each participant. All samples were positive for HBsAg and anti-HDV antibody. This was confirmed by ELISA (HDV Ab ELISA kit, Delaware Biotech, USA).
3.1. RNA Extraction and cDNA Synthesis
One hundred and forty microliter of sera was collected for RNA extraction using the QIAamp viral RNA mini kit (Qiagen, USA) according to the manufacturer's recommended procedures. Extracted RNA was eluted in 60 µL of elusion buffer. cDNA synthesis was performed with an Omniscript RT kit (Qiagen, USA) according to the manufacturer's instructions. HDV genotyping HDV genotyping was performed using a semi-nested PCR method. D120 forward primer (5'-ATGCCATGCCGACCCGAAGAGGAA-3'), DH1 (5'-GGCCTCTCAGGGGAGGATTCAC-3'), and D118 (5'-CTCAGGGGAGGATTCACCGACA-3') reverse primers were used in the semi-nested PCR, as described by Mirshafiee et al. (12). Briefly, for the first round of PCR amplification, the mixture contained 10 pmoL of each D120 and DH1 primers, and 10 mM dNTP and 5U Taq DNA polymerase (Cinna Gen, Iran). For the second round of PCR, 2 µL of the first-round PCR products was added to 10 pm of each D120 and D118 primers, and 10 mM dNTP and 5U Taq DNA Polymerase. PCR products were subjected for restriction enzyme digestion using SmaI and XhoI restriction endonucleases.
4. Results
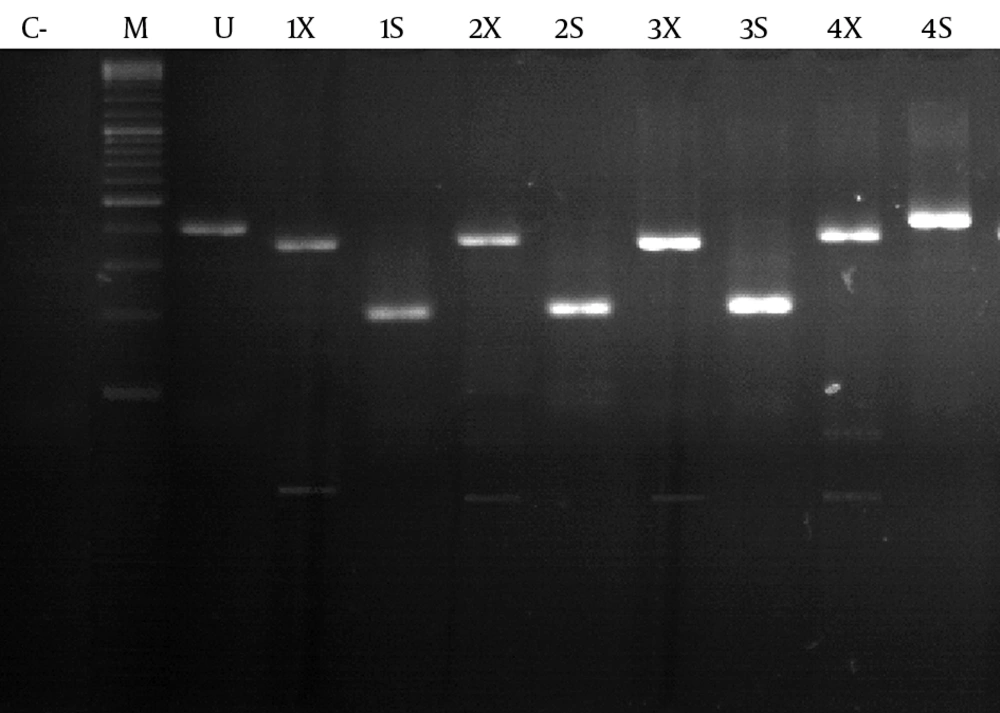
Of the 25 subjects, 9 were female and 16 were male. The mean age was 47 ± 10.14. In total, 12 cases (48%) were positive for HDV RNA. Of these 12 patients, 4 were female and 8 were male. All 12 patients were living in Mashhad when the study was performed; however, three patients were born in Afghanistan. At least 4 of the 12 patients had a family history of HDV infection. For HDV genotype 1, digestion with SmaI produced fragments of 219 and 222 bp. While with XhoI, fragments of 382 and 59 bp were produced (Figure 1). SmaI does not have any restriction site for the amplified fragments of HDV genotype 2. XhoI produces three fragments of 81 bp, 301 bp, and 59 bp (Figure 1). Genotype analysis of HDV RNA showed that the prevalence of HDV genotype 1 and 2 was 83.3% (n = 10) and 16.7% (n = 2), respectively.
5. Discussion
The genotyping of HDV could be performed using restriction fragment length polymorphism of PCR products (17), sequencing, and staining of liver biopsies with genotype-specific antibodies (18). In the Middle East, the predominant genotypes of HBV and HDV are D and 1, respectively. The association of HDV genotype 1 with HBV genotype D was reported in Pakistan, Turkey, Egypt, and Italy (19-21). Genotype 1 is reported to be the most prevalent genotype in different parts of Iran (12, 22, 23). Mohebbi et al. indicated that 25 patients from different parts of Iran who were positive for HDV antibody were as follows: 22 (88%) were HDV RNA positive and their phylogenetic analysis showed that all of them were infected by genotype 1 (clade 1). (23). Mirshafiee et al. indicated that 35 patients with HBsAg and anti-HDV positivity in Tehran. Among them, 13 (38.46%) cases were positive for HDV-RNA. They performed RT-semi-nestd PCR and RFLP and found that all cases were of genotype 1 (12).
Khalafkhany et al. showed that 31 anti-HDV seropositive patients in Iran showed that 15 (48.4%) were positive for HDV RNA and all belonged to genotype 1 (24). Regarding the neighboring countries of Iran, Perveen et al. performed indicated that in Karachi (Pakistan) that 22 patients all belonged to genotype 1 of HDV (25). In Turkey, HDV1 infection was reported to be endemic among HBsAg carriers, especially in southeastern Turkey. Phylogenetic analysis in 9 Turkish patients who were positive for anti-HDV showed that they were infected with genotype 1 (26). In other countries, analyses of HDV genotypes from 29 infected patients living in Ivaniushina showed a frequency of 48.27% and 51.72% for HDV genotypes 1 and 2, respectively (27). Also, Neverov et al. showed there was based on the phylogenic analysis of 66 patients in Russia and showed the same frequency (50%) for genotypes 1 and 2 (28).
A unique HDV genotype 2 in the Miyako Islands was first introduced for 6 patients with HDV-related chronic liver disease. This genotype showed low homology (75–81%) to the HDV genotype 2 reported from Japan but had a high identity (83–95%) with the novel genotype 2 (HDV genotype 2b), which was recently reported from Taiwan (29). A study in the Amazon region of Colombia on HBV and HDV sequences from 7 patients with fulminant hepatitis showed that five were positive for HBV-DNA and HDV-RNA. Of these 7 patients, 5 patients were positive for HDV genotype 3 (30). Among 233 HBsAg positive subjects in Cameroon, anti-HDV antibody was found in 17.6%. In addition, phylogenetic analyses showed the presence of HDV clades 1, 5, 6, and 7 (3). Barros et al. evaluated the seroprevalence of HDV among HBsAg chronic carriers in the Northeast Brazil. Among the studied 133 patients, 5 were positive for anti-HDV, of whom 3 were HDV RNA positive. HDV Clades 3 and 8 were found in 1 and 2 patients, respectively (31). Up to now, the epidemiology of HDV has not been comprehensively studied in Iran and limited data is available with regard to its genotypes, clades, and subgroups distribution.
This study was performed for the first time in Mashhad. Genotype analysis of HDV RNA showed that the prevalence of HDV genotypes 1 and 2 was 83.3% (n = 10) and 16.7% (n = 2), respectively. In addition, the findings indicate that the distribution of genotype 2 is not restricted to Taiwan, Japan, and the Miyako Islands and has spread over the Northern Asia in Russia. It is of interest that the two patients infected with HDV genotype 2 in the current study were not from Taiwan or Japan, where this genotype is more prevalent, and had not traveled to any of these countries, although, one of them had traveled to Saudi Arabia. This finding contradicts most studies carried out in Iran and in neighboring countries to Iran including Turkey and Pakistan, which showed no evidence of HDV genotype 2. These two patients were born and lived in Khorasan Razavi province (the province that includes Mashhad).
It is assumed that HDV infection in these regions has a homogenous origin or has become almost similar because of migration. The number of participants was not enough and further studies in this region with larger sample sizes are essential. In addition, the author’s encourage further studies in this region to determine the frequency of different HDV clades. Genotyping HDV helps us to determine its molecular epidemiology, geographical distribution, and identification of transmission chains.