1. Background
Halomethans are a group of methane derivatives that have substituent of halogen atoms (Cl, Br, I, or F) in methane molecule. Among the halogenated methanes, chloro-, bromo-, and chlorofluoro-methanes (Freon) are important industrial chemical compounds, widely used as solvent, refrigerant, and intermediates in chemical synthesis. Industrial productions of these compounds are about 2 million tons per year. Microorganisms naturally produce small amount of chlorinated methanes but fungi and a variety of marine algae produce an estimated amount of 5 million tons per year (1, 2). Halomethanes are chemically stable compounds and in different rates are soluble in water. Most of them are volatile and involved in degradation of the ozone layer. For example, dichloromethane lifetime in water and atmosphere are 700 years and 70 days, respectively. These compounds are highly toxic, mutagenic, and carcinogenic. Therefore, halomethanes can be considered as important environmental pollutants (3).
Biodegradation is the most important way to remove these compounds from the environment. Several aerobic methylotrophic bacteria and some of anaerobic bacteria that are able to degrade these compounds, including Hyphomicrobium, Methylobacterium, Methylophilus, Methanobacterium, Methanosarcina, and so on have been isolated (2, 4). Some bacteria can use halomethanes as their sole carbon and energy source, especially chloro- and dichloromethane but compounds such as tri- and tetra-chloromethane cannot be easily degraded and hence are cometabolized with methanol, methylamine, or formate. Methylotrophs are the most important chlorinated methane-utilizing bacteria.
Formaldehyde is the first product of degradation pathway in these bacteria that can be produced in periplasm by a methanol dehydrogenase (maxF) or in the cytosol by dichloromethane dehalogenase (dcmA) activities. Two parallel formaldehyde utilization pathways have been identified. In these pathways, formaldehyde reacts with tetrahydrofolate (THF) or tetrahydromethanoptrin (THMP) and methyl group is oxidized to CO2 through formate or incorporated in biosynthetic process and cell growth via the serine cycle. In some bacteria that are grown on halomethanes, methyl halide transferase is the key enzyme, which removes halogen, transfers the methyl group to THF, and then enters to the above-mentioned pathway (1, 2, 5).
In drinking water treatment process, some of disinfection byproducts can be formed during the reaction of disinfectants such as chlorine and bromide, with natural organic materials present in the water. The major halogenated disinfection byproducts that are commonly resulted from chlorine treatment are trihalomethanes (such as chloroform), haloacetic acids and so on (6, 7). Prolonged exposure to these chemicals in drinking water can cause some problems like cancer. There is some evidence of an association between chlorination of drinking water and increased risk of bladder, kidney, brain, and colorectal cancers (8). Hence, methylotrophic bacteria can use these compounds for growth; therefore, the presence of these bacteria in drinking water confirms the existence of these byproducts in chlorine treated drinking water. Halomethanes are volatile and working on them is difficult. Because of the ability of halomethane utilizing bacteria to grow on methanol, the appropriate method for primary isolation of these bacteria is culture on methanol containing media. Bacteria that are isolated in these media are methylotrophs and can be cultured on halomethane to study their growth and ability in biodegradation.
2. Objectives
This study aimed to represent a simple and rapid method for quantitative study of halomethanes utilizing bacteria in drinking water. Also, as natural biodegradation of chlorinated methanes is a very slow process, treatment by nanosilver was studied as a method to facilitate the biodegradation of these compounds in environment instead of using cometabolism.
3. Materials and Methods
3.1. Estimation of Methylotrophs Number by Most Probable Number
To study the presence of methylotrophs and most probable number (MPN) of them, drinking water samples were cultured in mineral salts medium containing: KH2PO4, 1.36 g; Na2HPO4, 2.13 g; MgSO47H2O, 0.3 g; (NH4) 2SO4, 0.5 g; CaCl22H2O, 1.99 mg; FeSO47H2O, 1.0 mg; MnSO4H2O, 0.35 mg; Na2MoO42H2O, 0.5 mg in 1000 mL distilled water with pH, 7.2 and up to 0.5% V/V (0.12 mol/L) of methanol (Merck, Germany) (9, 10). As the number of bacteria in the samples of treated drinking water is low, 3 L of water was first filtered by 0.45 µm pore size filter (Millipore, USA). Then, to separate the bacteria from the filter surface, it was transferred into a sterile Erlenmeyer flask and washed in 100 mL sterile distilled water with shaking for 30 minutes. Fifteen tubes of the medium were prepared and each set of 5 tubes inoculated with 10, 1, and 0.1 mL of water, which the filter was washed in. To prevent evaporation of alcohol from the medium, tubes were sealed with parafilm and incubated at 30ºC for 20 days. Later, the tubes with turbidity or pellets (indicative of bacterial growth) were selected for isolation of methylotroph bacteria and subsequent experiments.
3.2. Assessment of Halomethane Utilizing Bacteria
MPN tubes that showed turbidity and bacterial growth were used for assessment of halomethane utilizing bacteria. As chloromethane is in gas form and its application was difficult, also, tri- and tetra-chloromethanes are less biodegradable than the others, the dichloromethane, which is a liquid form of the chlorinated methane compounds was used as an ideal compound in determination of chlorinated methane utilizing bacteria. Fifty microliter of dichloromethane (Merck, Germany) was added to each tube. Because of the volatility of this compound the tubes were sealed with a sterile rubber cap. Then, they were incubated for 72 hours at 30ºC (10). An uncultured tube containing 50 µL of dichloromethane was used as control. The amount of released chloride ion in the medium was measured after 72 hours by Istek pH/ISE meter (pH-240L model, Korea). The electrode of chloride ion was able to measure 1.8-35500 ppm of chloride ion concentrations. NaCl standard solutions of 10, 100, and 1000 ppm were used to calibrate pH/ISE meter. Then, chloride ion concentration was determined in each sample. The difference of chloride content between the test groups and the control was calculated. This value represents the amount of released chloride ion by halomethane utilizing methylotroph bacteria in drinking water sample.
3.3. Isolation and Identification of Methylotrophic Bacteria
Mineral salt medium containing 0.6% (V/V) of methanol and 1.5% ultra-pure agar was used for isolation and purification of methylotrophic bacteria present in positive MPN tubes. The selected bacteria were identified by Gram staining and standard biochemical tests according to Bergey’s Manual of Determinative Bacteriology.
3.4. Study of Nanosilver Effect on Chloroform
Nanosilver effect on chloroform (trichloromethane) was studied by two methods (spectrophotometry and Hantzsch reaction). For evaluation by spectrophotometry method, nanosilver 0.7 ppm (Nanocide Company, Iran) was added to 1 mL of chloroform (Merck, Germany) and after 1 and 5 minutes absorbance was read at 293 nm wavelength. One milliliter of water, with 0.7 ppm nanosilver was used as control. For evaluation by Hantzsch reaction, 1 mL of chloroform was treated with 0.7 ppm of nanosilver and then 1 mL of reagent (ammonium acetate 2 M, acetic acid 0.05 M, acetyl-acetone 0.02 M) was added and incubated at 60ºC for 10 minutes. The yellow color indicated the presence of formaldehyde (11).
3.5. Effect of Nanosilver on the Growth of Isolated Bacteria in MPN Test in Chloroform
One hundred and twenty microliter of bacterial suspension (1 McFarland concentration) was inoculated into 3 mL of Bushnell-Haas Broth (Himedia, India) containing 1% and 0.1% of chloroform treated with nanosilver 0.2 ppm. Two chloroform samples of 1 mL and 0.1 mL in medium were used as controls. Then, the tubes were sealed with a rubber cap. After one week incubation at 30ºC, optical density (OD) of the bacteria was measured to obtain growth rate.
3.6. Formic Acid Production by the Isolated Bacteria
In this method, bacterial cultures were prepared as mentioned above and after 48 hours of incubation at 30ºC centrifuged for 20 min at 1000 ×g. Then, a few drops of phenol red were added to 1 mL of the supernatant. Color changes from red to yellow indicated acid production by the bacteria. Fresh culture medium with and without formic acid solution were used as positive and negative controls, respectively.
3.7. Evaluation of Chloroform Utilization in the Isolated Bacteria by Gas Chromatography
One hundred microliter of bacterial suspension with OD = 0.4 was inoculated in 3 mL of Bushnell-Haas Broth containing 3 µL of chloroform and also in 3 mL of Bushnell-Haas Broth containing 3 µL nanosilver treated chloroform. The same tubes, without the bacteria, were used as controls. All tubes were incubated at room temperature with shaking at 120 rpm for 48 hours. Samples were analyzed using an Agilent 6890 Gas Chromatograph (USA) (5, 12). One microliter of headspace gas of each tube was injected onto head of chromatographic column (HP-1). Initial temperature of column was 40ºC and increased to final temperature of 100ºC with the rate of 3ºC per minute. Detector temperature was 220ºC.
4. Results
Detection of methylotrophs and probable number in water by MPN method: In MPN test, 6 out of 15 tubes were positive. Turbidity or pellet formation patterns were 5-1-0 of 3 sets of 15 tubes, respectively. According to MPN table and volume of filtered water most probable number of methylotrophs in tested drinking water was 10 MPN Index/L. Assessment of halomethane utilizing bacteria, isolation, and identification of them: The results of dichloromethane degradation in 6 positive tubes of MPN test are shown in Table 1. Out of these 6 tubes, which contained methylotroph bacteria, the released chloride ion in 4 tubes was significant, indicative of the presence of dichloromethane utilizing bacteria. The patterns of these 4 positive tubes were 3-1-0 of 15. According to MPN table and volume of filtered water most probable number of halomethane utilizing methylotrophs in drinking water sample was 4 MPN Index/L.
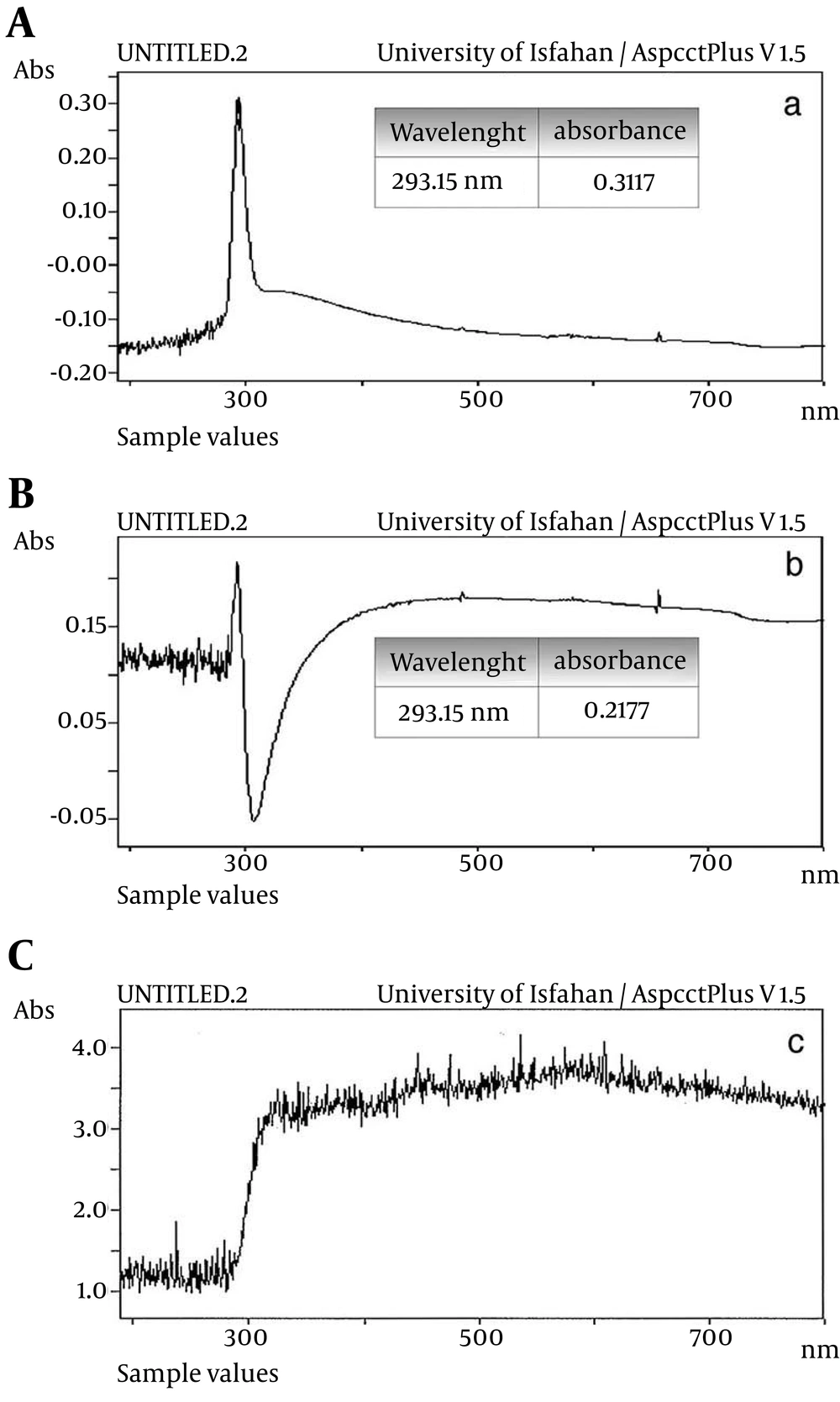
Seven bacteria were isolated from positive MPN tubes. One of them, which had the highest growth in methylotrophic condition and identified as Xanthomonas was selected for the study of nanosilver effect on chloroform biodegradation. Effect of nanosilver on chloroform, bacterial growth (chloroform utilization) and formic acid production: As it is shown in Figure 1, the absorbance was decreased from 0.31 to 0.21 (about 33%) after one minute (Figure 1A, 1B). This indicated the hydrolysis of chloroform. The hydrolysis process was completed after 5 minutes (Figure 1C). In Hantzsch reaction the production of formaldehyde was confirmed by the appearance of yellow color.
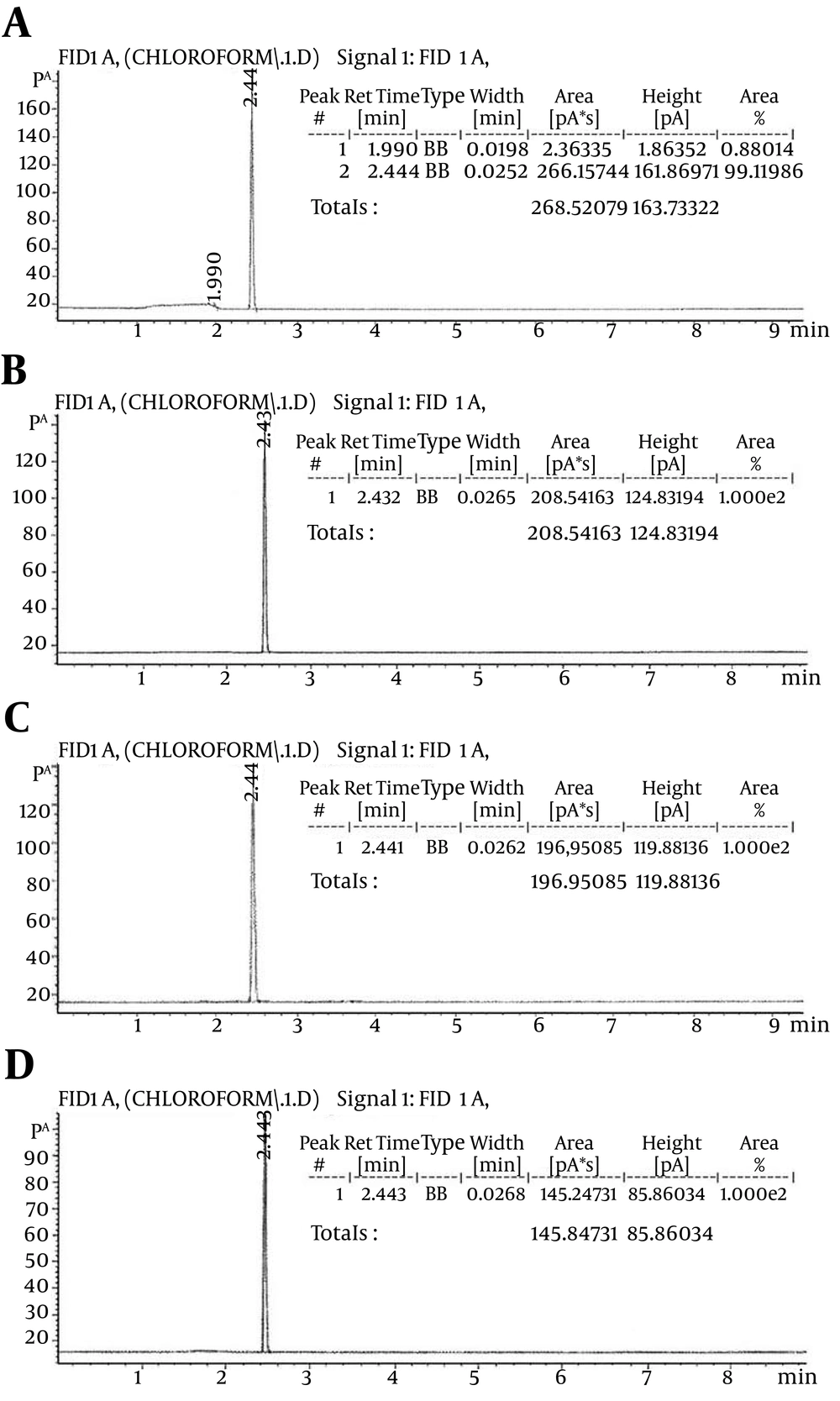
The highest growth and formic acid production (color changes) were observed in tubes containing 1% chloroform, treated with nanosilver, and then the tubes containing 0.1% chloroform, treated with nanosilver, and 1% and 0.1% non-treated chloroform were followed, respectively. Evaluation of chloroform degradation by Gas Chromatography method (GC): As it is shown in Figure 2A, the highest amount of chloroform was detected in the medium containing the chloroform without bacteria (area = 266.15). The calculated area in the medium containing the chloroform, which treated with nanosilver without bacterium and medium containing the chloroform with the bacteria were 208.54 and 196.95, respectively (Figure 2B, 2C). The lowest amount (area = 145.54) was observed in the medium that contained the bacterium and chloroform treated with nanosilver (Figure 2D).
| MPN tubes with growth (containing methylotrophs) | Chloride content, mg/L | Released Chloride, mg/L |
|---|---|---|
| 1 | 160 | 44 |
| 2 | 178 | 62 |
| 3 | 173 | 57 |
| 4 | 117 | 1 |
| 5 | 118 | 2 |
| 6 | 169 | 53 |
a Chloride ion concentration in control tube was 116 mg/L.
A, Medium containing the chloroform without bacterium (area = 268.52); B, medium containing the chloroform that treated with nanosilver (area = 208.54); C, medium containing the chloroform and bacterium (area = 196.95); D, medium containing the chloroform that treated with nanosilver and bacterium (area = 145.84).
5. Discussion
Chlorinated methanes are important industrial chemicals as well as significant environmental pollutants. Several reports about the biodegradation of chloromethane and dichloromethane by bacteria have been published (1, 2, 12). There are a few reports about other chlorinated compounds such as trichloromethane (chloroform) and tetrachloromethane (4, 13). Although the biodegradation of mono- and di-chloro compounds by bacteria is easier than tri- and tetra-chloro ones and many bacteria can use these as carbon and energy source, biodegradation of tri- and tetra-chlorinated chemicals occurs by cometabolism with other compounds such as methane and butane (14, 15). The two goals of this study were representing a simple method for quantitative study of chloromethanes utilizing bacteria in water and another method for facilitating the biodegradation of these compounds in a way other than cometabolism.
First, to represent a rapid and simple method for quantitative study of chloromethanes utilizing bacteria in drinking water, the presence of methylotroph bacteria confirmed by MPN method on methanol containing medium. Then dichloromethane was added to the tubes with the grown bacteria and after incubation, its biodegradation was determined by measuring the released chloride ions. Thus, we combined two tests to introduce a rapid method for determination of halomethane utilizing bacteria and estimation of the most probable number of them in drinking water.
Second, the effect of nanosilver on biodegradation of multiple chlorinated methanes (such as chloroform) as sole carbon source by bacteria was studied. Methylotrophs and mono- and di-chlorinated methane consuming bacteria convert these compounds to formaldehyde at the first step of metabolic pathway and then formic acid is produced. Final product is CO2. Halomethanes that have more halogenic groups such as tri- and tetra-chloromethane do not enter in the mentioned metabolic pathway, and degradation pathways of them are not well understood. However, if these compounds are converted to the formaldehyde, they will be utilizable for the bacteria. This also can be done by treating of the compounds with nanosilver. As the spectrophotometric absorbance, formaldehyde production, acid formic production, and GC tests showed, chloroform treatment by nanosilver leads to dechlorination and formaldehyde production. In this way, the degradation of these chlorinated compounds by bacteria will be easier and faster. As a result, the degradation of these chemicals is more efficient without the need for cometabolism. It has already been reported that nanosilver is bactericidal in certain concentrations (16), but according to the results obtained in this study, 0.2 ppm concentration of nanosilver not only had any antibacterial effects, but also increased the activity of bacteria on chloroform biodegradation. Hence, this method can be used for biological removal of these environmental pollutants.