1. Background
Enterococci are Gram-positive cocci that have been classified as group D streptococci. Enterococci are members of the normal flora of animals and humans (1). Enterococci have both an intrinsic and acquired resistance to antibiotics, making them important nosocomial pathogens. Increasing antimicrobial resistance among pathogens that cause nosocomial infections constitutes a major public health problem in the worldwide (2). Vancomycin has been used in human medicine since 1958 (3). Vancomycin resistant Enterococcus species were initially reported in 1988 (2, 4). Since then, Vancomycin Resistance Enterococci (VRE) has made a serious clinical problem and spreads in hospitals of many countries (5). According to studies that have been done around the world, we need to determine the prevalence of VRE and the dominant genotype in Arak. Vancomycin resistance enterococci can remain viable in the environment for an extended period of time and therefore poise a problem for infection control in hospitals; this has made enterococci the second leading cause of nosocomial infections in the world (6).
Enterococci have been detected as part of the enteric flora in non-symptomatic patients. These patients are potential sources for transfer of this organism to other patients and medical personnel. Use of vancomycin may also increase VRE in patients by eliminating other colonizing bacteria and allowing the development of VRE (7). Enterococci possess the capacity to share antibiotic resistance genes among themselves and other pathogenic bacteria such as Staphylococcus and Streptococcus species. Six different types of vancomycin resistance have been shown by Enterococcus species including Van-A, Van-B, Van-C, Van-D, Van-E and Van-F. In Europe, VanA-type Enterococcus faecium is the predominant type isolated from animals, humans, and environmental sources. The VanA gene is encoded on the Tn1546 transposon or a Tn1546-like transposon (8) and the VanB gene is encoded on a conjugative transposon, Tn1549 (9), and may potentially be associated with the transfer of resistance to other organisms, including Staphylococcus aureus (10). VanC-type glycopeptide resistance is constitutive; therefore, transfer to other organisms is not of much concern (11).
2. Objectives
The aim of the present study was to monitor and detect resistance genes in VRE isolated from hospitalized patients. This paper presents the findings of this research over a 12-month period.
3. Materials and Methods
3.1. Bacterial Isolates
Two hundred randomly selected enterococci were isolated, during a 12-month period (April 2010 to April 2011), from clinical specimens (urine, blood, wound, sputum and catheter) available at Arak educational hospitals, Iran. Enterococcus faecium, Enterococcus faecalis V583, Enterococcus casseliflavus ATCC 25788, and Enterococcus gallinarum GS were used as controls (7).
3.2. Identification and Antimicrobial Susceptibility Testing of Enterococcus Isolates
Isolated enterococci were identified by 6.5% NaCl tolerance and growth on bile-esculin agar (Merck, Germany) with esculin hydrolysis. Two hundred enterococci samples collected from clinical specimens were tested for susceptibility to tetracycline (30 μg, Himedia, India), gentamicin (30 μg, Himedia, India), erythromycin (30 μg, Himedia, India), teicoplanin (30 μg, Himedia, India), co-trimoxazole (30 μg, Himedia, India), ciprofloxacin (30 μg, Himedia, India), linezolid (30 μg, Himedia, India) and vancomycin (30 μg, Mast, Germany) by an agar incorporation method in accordance with the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (12). Discs containing specific concentrations of antibiotics were placed on solid media containing the bacteria (with the bacteria concentration equivalent to 0.5 McFarland standard). The antibiotic was released into the culture medium to prevent the growth of bacteria. By measuring the circle created around the discs, the resistance or susceptibility of bacteria was determined. Micro dilution test (MICs) for vancomycin and teicoplanin was carried out in sterile microdilution plates with 96 flat-bottomed wells, composed of certain amounts of Muller-Hinton broth (2x) and antibiotics (2, 4, 8, 16, 32, 64, 128, and 256 µg/mL). Next, isolates at the concentration of 5 × 105 CFU/mL were added to each well. The MICs were determined after 24 hours of incubation at 37°C (12).
3.3. Vancomycin Resistance Gene Amplification by the Polymerase Chain Reaction
Chromosomal DNA was prepared according to the standard CTAB/NaCl method. Briefly, after the pellet of bacterial culture was resuspended in Tris-EDTA buffer (Tris 10 mM, EDTA 1 mM, pH 8), the bacteria were lysed by sodium dodecyl sulfate (SDS) and proteinase K. The chromosomal DNA was extracted by CTAB/NaCl solution (10% CTAB and 0.7 M NaCl). The cell debris and proteins were removed by two times phenol/chloroform/isoamyl alcohol (25: 24: 1) mixture. Next, DNA was precipitated by isopropanol and washed in ethanol (70%), dried, and then resuspended in TE buffer. The PCR reaction mixture and conditions for amplification of the VanA were as follows; 95°C for five minutes followed by 31 cycles of denaturation at 95°C for one minute, annealing at 51°C for one minute, extension at 72°C for one minute and final extension at 72°C for five minutes.
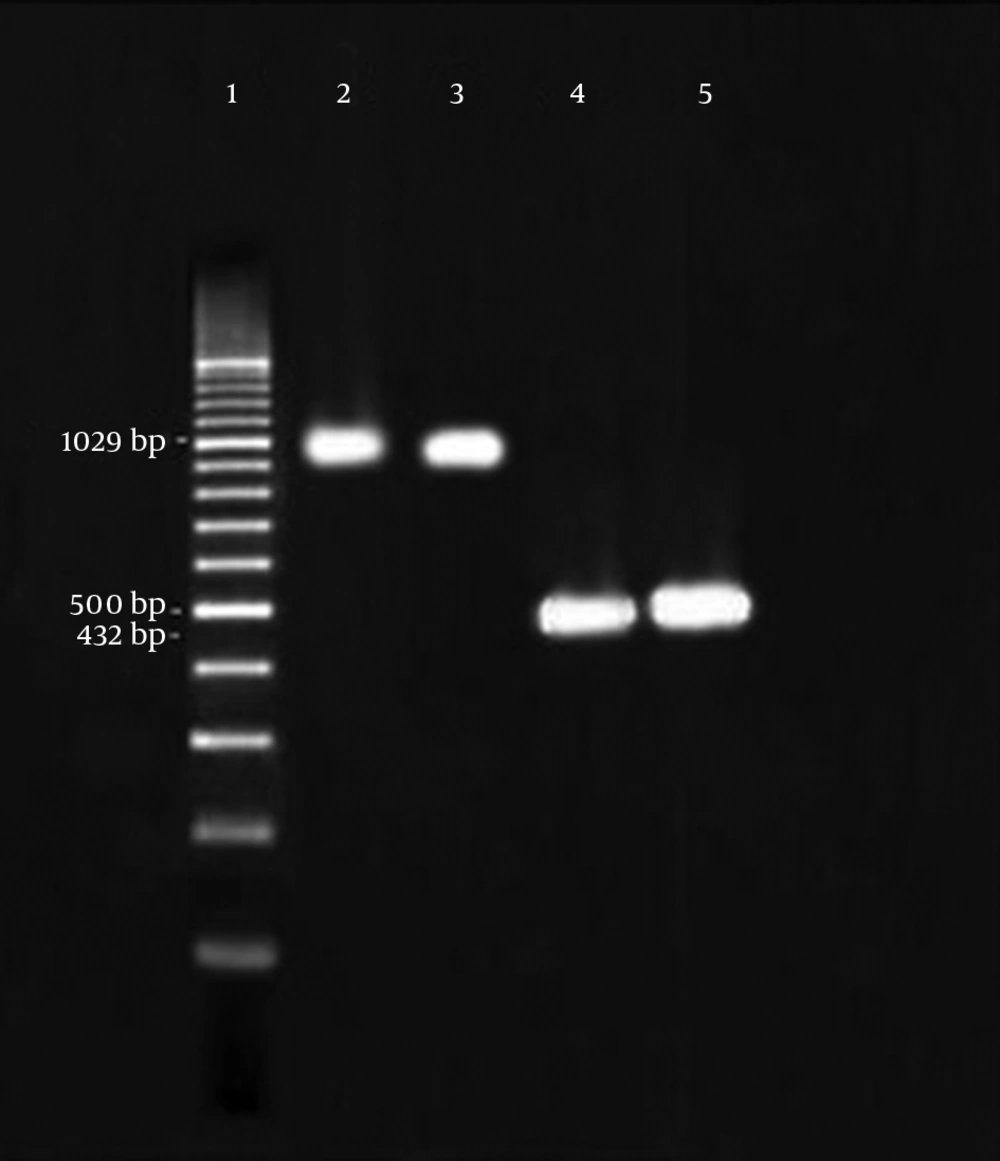
The reactions were initiated in a solution containing 200 μM concentrations of dNTPs (Cinagen, Iran), 10 pM of each primer, 50 mM of MgCl2 (Cinagen, Iran), 0.5 U of Taq polymerase (Cinagen, Iran) and 0.5 μL of DNA template in a final volume of 25 μL. Amplification of the VanB was performed at 95°C for five minutes followed by 30 cycles of denaturation at 95°C for one minute, annealing at 57°C for one minute, extension at 72°C for one minute and final extension at 72°C for five minutes. Furthermore, amplifications of the VanC1 and VanC2/3 were performed at 95°C for five minutes followed by 36 cycles of denaturation at 95°C for one minute, annealing at 54°C for one minute, extension at 72°C for one minute and final extension at 72°C for five minutes. The PCR products were analyzed by electrophoresis on horizontal 1% agarose gels in TBE 1X buffer loaded with 5 μL of reaction mixture and stained with ethidium bromide after electrophoresis (Figure 1). The primers and fragment sizes are listed in Table 1.
| Gene | Oligonucleotide Sequence, 5ʹ-3ʹ | PCR Product Size, bp | References |
|---|---|---|---|
| VanA | 885 | (13) | |
| FvanA | CATGAATAGAATAAAAGTTGCAATA | ||
| Rvan | CCCCTTTAACGCTAATACGATCAA | ||
| VanB | 885 | (13) | |
| FvanB | GTGACAAACCGGAGGCGAGGA | ||
| RvanB | CCGCCATCCTCCTGCAAAAAA | ||
| VanC1 | 467 | (14) | |
| FvanC1 | GGTATCAAGGAAACCTC | ||
| RvanC1 | CTTCCGCCATCATAGCT | ||
| VanC2 | 429 | (15) | |
| FvanC2/C3 | CGGGGAAGATGGCAGTAT | ||
| RvanC2/C3 | CGCAGGGACGGTGATTTT |
4. Results
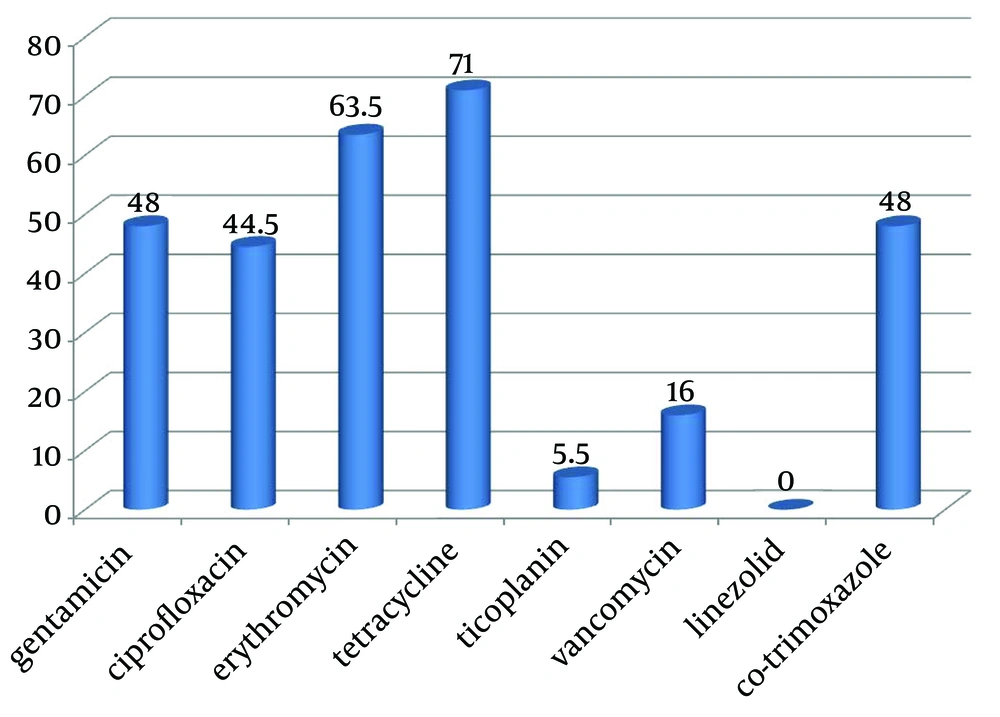
From April 2010 to April 2011, a total of 200 positive samples for enterococci were processed. Among the 200 strains, 164 (82%) were isolated from urine, 16 (8%) from wound, nine (4.5%) from sputum, eight (4%) from blood and three (1.5%) from catheters. Disk diffusion agar showed that 96 strains were resistant to gentamicin, 89 to ciprofloxacin, 127 to erythromycin, 142 to tetracycline, 11 to teicoplanin, 32 to vancomycin, none to linezolid and 96 to co-trimoxazole (Figure 2). The MIC test was performed to distinguish intermediate and resistance strains to vancomycin. Minimum Inhibitory Concentrations illustrated that, 88 strains had MIC of ≥ 32 μg/mL to vancomycin and 59 strains had MIC of ≥ 32 μg/mL to teicoplanin.
4.1. Molecular Analysis
Molecular studies detected that 59.09% of VRE contained the VanA genes and 7.95% of vancomycin resistant enterococci contained the VanB gene (Figure 2). None of the strains had the VanC1 and VanC2/3 genes.
5. Discussion
Enterococcus is known as an important pathogen in Iran like other parts of the world. The increasing use of vancomycin makes VRE important nosocomial pathogens. Vancomycin in combination with an aminoglycoside can provide effective treatment for severe Enterococcus infections, while resistance to vancomycin antibiotic is increasing in enterococci. In this study, the pattern of antibiotic resistance and prevalence of vancomycin resistance in enterococci was explored. Antibiotic resistance is a serious and growing phenomenon in contemporary medicine and has emerged as one of the pre-eminent public health concerns of the 21st century, in particular as it pertains to pathogenic organisms. In this study, 200 different strains of enterococci were isolated from hospitalized patients and outpatients.
Overall, 82% of these Enterococcus strains were isolated from urine samples; therefore the role of enterococci in urinary tract infections was further illustrated. Similarly, Ranjbar et al. (16) showed that enterococci were isolated from 8.7% of urine samples from the Children's Medical Center of Tehran, and were the fourth cause of urinary tract infections. Also the results of this study showed that among the isolated VRE, there were high levels of resistance to most of the tested antimicrobial agents. The findings also revealed that all the strains were sensitive to linezolid. Thus, our results showed that infections caused by multiple antibiotic-resistant enterococci are common, and this is a serious health threat as treatment of these strains is very difficult and controlling the spread of these microorganisms is important. In addition, lack of routine screening for VRE strains and multi-resistant strains might be a reason for the increasing resistance rate to vancomycin in enterococci, isolated in the current study. Thus restricted use of antibiotics, based on the results of the antibiotic sensitivity patterns, is recommended.
Vancomycin is the drug of choice for infections caused by multi-drug resistant enterococci. Multi-drug resistance is commonly seen in people who have recently been treated with antibiotics (17). In the present study, 59% of vancomycin-resistant strains carried the VanA gene and 7.95% of VRE strains carried the VanB gene. In epidemiological studies from different parts of Iran, VanA gene was more prevalent than VanB gene. It should be mentioned that our findings were compatible with the studies of other researchers. According to their results, amongst 54 enterococci isolated from clinical samples, the least resistance was observed to linezolid. Furthermore, 69% of resistant strains contained VanA (15). Teymourrnejad et al (18) studied 422 strains of enterococci and found that 60% of the isolates contained VanA, 40% vanB and 20% had both VanA and VanB (19). Our results were similar to those of other Asian studies. In 2011, Xu et al. (13) obtained 32 VRE strains from a tertiary-care hospital of Beijing, china. All the isolates harbored the VanA gene; however, four exhibited the VanB phenotype (13). Other studies have shown that, except for VanA and VanB genes, the other vancomycin resistance genes are in very low abundance in Iran (18, 20). Kirdar et al. (14) showed that all 12 E. faecium were isolated from the hematology unit. They detected VanA gene by PCR yet VanB and VanC genes were not found in the 12 VRE strains (14).
In conclusion, the rates of VRE in this study were notable. Since glycopeptide antibiotics, such as vancomycin and teicoplanin, are the drugs of choice and are often the last options for treatment of hospital infections caused by multiple drug resistant Gram-positive bacteria, thus antibiotic resistance in enterococci against glycopeptide antibiotics is considered as a threat to nosocomial infections. One concern is that VRE appear to be able to transfer vancomycin resistance to unrelated bacteria such as methicillin resistant Staphylococcus aureus (MRSA). In addition, VRE organisms are usually resistant to more than one antibiotic. Vancomycin Resistance Enterococci can also spread from person to person and are an increasing problem in hospitals and chronic-care facilities. To avoid spreading of VRE from person to person, it is important to wash or decontaminate hands frequently, including before and after touching the patient or his/her environment. In the hospital, staff should also wear gowns and gloves when caring for a person with VRE.