1. Background
Pseudomonas aeruginosa is an important opportunistic pathogen with the ability to propagate on medical devices, hospital environments and even in disinfectants (1). Due to the intrinsic resistance to many antimicrobial agents, P. aeruginosa infections present serious therapeutic challenges both in the community and health centers. In addition, the organism can acquire resistance elements to multiple classes of antibacterial agents, even during the course of treatment (2, 3). P. aeruginosa isolates from burn wounds are often responsible for systemic sepsis, graft loss, prolonged hospital stay and mortality (4).
Several mechanisms are responsible for resistance to β-lactam antibiotics in P. aeruginosa including: genetic mutations that lead to stable over expression of chromosome-mediated AmpC cephalosporinases, acquisition of transferable β-lactamase genes, overproduction of efflux systems, and reduced permeability (5). Among the numerous β-lactamases, Ambler class A extended-spectrum β-lactamases (ESBLs) and class B metallo β-lactamases (MBLs) are reported as rapidly growing enzymes in clinical isolates of P. aeruginosa. ESBLs are capable of hydrolyzing penicillins, cephalosporins, and aztreonam (except for cephamycins or carbapenems). These enzymes are inhibited by β-lactamase inhibitors such as clavulanic acid (6, 7). Among more than 200 different known ESBLs in Gram-negative bacteria, 32 are detected in P. aeruginosa belonging to class A (TEM, SHV, CTX-M, PER, VEB, GES, BEL), and class D (OXA type) β-lactamases (8).
AmpC β-lactamases hydrolyze cephamycins (e.g. cefoxitin and cefotetan), oxyimino-cephalosporins (e.g. ceftazidime, cefotaxime, and ceftriaxone), and monobactams (e.g. aztreonam) (9). In P. aeruginosa, decreased susceptibility to the extended-spectrum cephalosporins such as ceftazidime mostly results from over expression of the naturally occurring AmpC β-lactamases (10). Among the carbapenems, intermediate susceptibility to imipenem is widely associated with the modification of the outer membrane protein OprD, production of MBLs and over expression of the cephalosporinases associated with decreased outer membrane permeability. Recently, a novel resistance mechanism was shown in P. aeruginosa, where peculiar AmpC β-lactamases with expanded-spectrum activity toward imipenem may also contribute to decreased imipenem susceptibility (11).
2. Objectives
The current study aimed to determine the prevalence of ESBL and MBL production among AmpC β-lactamase producing P. aeruginosa isolated from burns and evaluate the presence of their related antibiotic resistance genes.
3. Materials and Methods
3.1. Bacterial Isolates
Fifty one non-duplicate P. aeruginosa isolates were randomly selected from a collection of burn isolates provided by Shahid Motahari Hospital in 2011. The antibiotic susceptibility patterns and MBL production were previously determined using disc diffusion and the double disc synergy tests, respectively (12). The isolates were maintained in brain heart infusion broth (Oxoid, UK) containing 10% dimethyl sulfoxide and stored at -20ºC until use. Controls for polymerase chain reaction (PCR) tests were: P. aeruginosa PAO1 containing blaAmpC (provided by Dr. Abdi, Al-Zahra University, Iran), DNA from Klebsiella pneumoniae 7881 harboring blaTEM, blaSHV and blaCTX-M, P. aeruginosa KOAS carrying blaPER-1 and P. aeruginosa 10.2 harboring blaVEB-1 were obtained from Pasteur Institute, Tehran, Iran. P. aeruginosa ATCC 27853 was also used as the susceptible control strain.
3.2. Antimicrobial Susceptibility Testing
Antibiotic susceptibility testing of the isolates was carried out by the disc diffusion method, according to the Clinical and Laboratory Standards Institute guidelines (CLSI) (5). The used antibiotic discs (MAST Group LTD, Merseyside, UK) were: ceftazidime (30 μg), aztreonam (30 μg), carbenicillin (100 μg), piperacillin (100 μg), ticarcillin (75 μg), cotrimoxazole (25 μg), amikacin (30 μg), cefepime (30 μg), ciprofloxacin (5 μg), tobramycin (10 μg), meropenem (10 μg), imipenem (10 μg), cefoxitin (30 μg), and piperacillin/tazobactam (100/10 μg).
3.3. Determination of Minimum Inhibitory Concentrations
Minimum inhibitory concentrations (MICs) for imipenem and ceftazidime were determined by the microdilution assay as recommended by the CLSI (13).
3.4. Phenotypic Detection of AmpC Production
Initial screening for AmpC β-lactamase production was performed by the AmpC disc test. The results showed a zone diameter of < 18 mm for cefoxitin (14). Briefly, a blank disc moistened with sterile saline, was inoculated with a few colonies of the test strain. The disc was then placed next to a cefoxitin disc (30 μg) on the surface of a Muller Hinton agar plate (Leofilchem, Italy) previously inoculated with a lawn of Escherichia coli ATCC 25922. The plate was incubated overnight at 37ºC. An indentation of the cefoxitin inhibition zone adjacent to the disc containing the test strain was an indication of AmpC β-lactamase production.
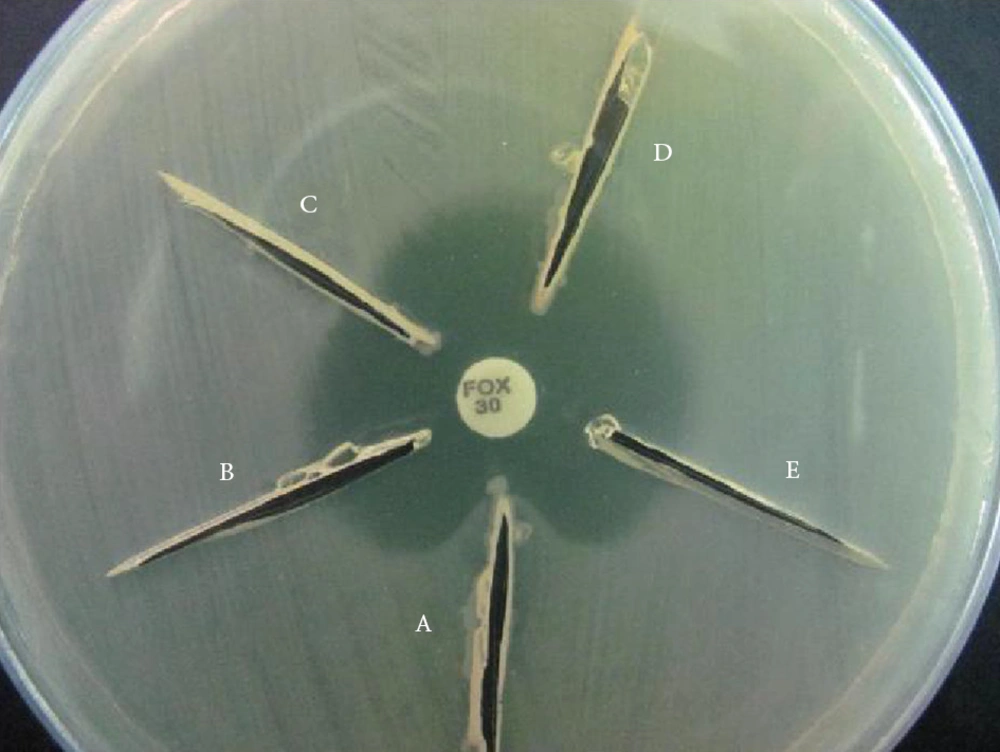
AmpC production was also determined using a modified three-dimensional extract test (TDET) (15). Briefly, bacterial colonies from overnight cultures on Mueller Hinton agar were transferred to a pre-weighed sterile micro centrifuge tube. The tube was weighed again to obtain 10 to 15 mg of bacterial wet weight per sample. The bacteria were then suspended in peptone water (Merck, Germany) and centrifuged at 3000 rpm (Sigma 1-13 microcentrifuge, Germany) for 15 minutes. Crude cell extracts were prepared by 10 cycles of alternate freeze-thawing at -78ºC (dry ice/ethanol bath) and 37ºC. The extracts were then used to screen for AmpC using the following method. Linear slits (3 cm) were cut into plates inoculated with each test organism using a sterile scalpel blade, 3mm away from a cefoxitin disc in an outward radial direction. Approximately 40 to 50 μL of the prepared extract was loaded into each slit and the plates were kept upright for 5 to 10 minutes followed by incubation at 37ºC overnight. AmpC production was recorded when clear distortion of cefoxitin inhibition zones were observed (Figure 1).
3.5. Phenotypic Detection of ESBL
ESBL production was detected by the conventional double disc synergy test (DDST) using ceftazidime (30 μg) and cefotaxime discs (30 μg) with or without clavulanic acid (10 μg) as recommended by the CLSI. An increase of ≥ 5 mm in the inhibition zones of either cephalosporin in combination with clavulanic acid compared to the cephalosporin alone was interpreted as ESBL production (13).
3.6. PCR Amplification of AmpC and ESBL Genes
DNA extraction was carried out by boiling. Briefly, a loopful of an overnight grown culture of each test isolate as well as the control strains were suspended in 500 μL of sterile double distilled water, boiled at 100ºC for 10 minutes and were centrifuged at 12’000 × g for 10 minutes. The supernatants were then used as DNA template for PCR amplification tests.
PCR amplifications were performed using specific primers for Ambler class A (blaTEM, blaSHV, blaCTX-M, blaVEB-1, blaPER-1) and class C (blaAmpC) β-lactamase genes (Table 1). AmpC amplifications were initially performed using a pair of external primers (PreAmpC-PA1 and PostAmpC-PA2) yielding a 1243-base pair product encompassing the entire AmpC gene of P. aeruginosa, but excluding its promoter sequences. Another pair of primers (AmpC-PA-A and AmpC-PA-B) was also used as internal primers for AmpC gene yielding a 1063 bp product (Table 1).
PCR experiments were carried out in 25 μL volume reaction mixtures containing 10 pM of each primer, 200μM dNTP, 1.4 mM MgCl2, 1 μL of crude DNA template and 1U of Taq DNA polymerase in the reaction buffer provided by the manufacturer (CinnaGen, Tehran, Iran). Amplifications were carried out in thermocycler (Techne, UK) using the following program: five minutes denaturation at 95ºC followed by 30 cycles of 95ºC for one minute, one minute at the annealing temperature (43 to 63ºC for ESBLs, 62ºC and/or 65ºC for AmpC), and one minute at 72ºC with a final extension period of 10 minutes at 72ºC.
3.7. Statistical Analyses
Comparisons of β-lactamase phenotypes and presence of the related genes were performed using the chi-square test. Correlation between antibiotic resistance and β-lactamase gene carriage was analyzed by the nonparametric Mann-Whitney U test. All statistical analyses were carried out using SPSS software version 19. A P Value of < 0.05 was considered statistically significant.
| β-lactamase (Primer) | Sequence (5' to 3') | Product Size (bp) | Reference |
|---|---|---|---|
| Class A | |||
| TEM-F | GAGTATTCAACATTTCCGTGTC | 851 | (10) |
| TEM-R | TAATCAGTGAGGCACCTATCTC | ||
| SHV-F | AAGATCCACTATCGCCAGCAG | 231 | (10) |
| SHV-R | ATTCAGTTCCGTTTCCCAGCGG | ||
| CTX-M-F | CGCTTTGCGATGTGCAG | 550 | (16) |
| CTX-M-R | ACCGCGATATCGTTGGT | ||
| PER-1-F | ATGAATGTCATTATAAAAGC | 920 | (10) |
| PER-1-R | AATTTGGGCTTAGGGCAGAA | ||
| VEB-1-F | CGACTTCCATTTCCCGATGC | 643 | (10) |
| VEB-1-R | GGACTCTGCAACAAATACGC | ||
| Class C | |||
| PreAmpC-PA1 | ATGCAGCCAACGACAAAGG | 1243 | (11) |
| PostAmpC-PA2 | CGCCCTCGCGAGCGCGCTTC | ||
| ampC-PA-A | CTTCCACACTGCTGTTCGCC | 1063 | (11) |
| ampC-PA-B | TTGGCCAGGATCACCAGTCC |
4. Results
Disc diffusion results showed that all test isolates were resistant to three or more antibiotic classes; therefore, they were identified as multidrug resistant (MDR). Antibiotic resistance rates shown in Figure 2 were: piperacilin, carbenicillin, tobramycin and cefoxitin (100%), imipenem, ticarcillin, aztreonam and cotrimoxazole (98%), amikacin, cefepime and ciprofloxacin (96%), meropenem (94%), ceftazidime (92%) and piperacillin/tazobactam (88%). The MIC results showed 88.2% resistance to ceftazidime and 98% to imipenem, confirming the results obtained by the disc diffusion method. There was no significant association between antibiotic resistance patterns and β-lactamase production. AmpC production was observed in 35 isolates (68.6%) using the AmpC disc test and only 18 (51.4%) by the TDET test suggesting the superiority of the AmpC disc test to TDET. ESBL production was observed in 20 isolates (39.2%) and 19 isolates (37.3%) were MBL producers. Of the 35 AmpC producers, two (5.7%) had the ESBL phenotype, 13 (37.1%) produced MBL and six (17.1%) were positive for all three enzymes.
PCR amplification results showed that 43 isolates (84.3%) carried β-lactamase genes. BlaAmpC was present in 31 isolates (60.8%), two of which were susceptible to ceftazidime (Table 2). There was a significant correlation between the AmpC disc test phenotype and AmpC gene carriage (P < 0.05), while no relationship was observed between TDET and presence of the AmpC gene. Among the class A β-lactamases, 20 isolates (39.2%) harbored blaTEM and 11 (21.6%) carried blaPER-1 genes. SHV, CTX-M and VEB-1 genes were not detected in any of the isolates. All MBL producing isolates harbored their related genes (12). Overall, there was correlation between β-lactamase productions with the presence of their related genes in the test isolates (P < 0.05).
a MIC (µg/ml) (NO. Susceptible) (MIC, Minimum Inhibitory Concentration)
b Extended-Spectrum β-Lactamase
c Metallo-β-Lactamase
5. Discussion
Multiple β-lactamase producing P. aeruginosa cause major therapeutic failure and pose significant clinical challenges if remain undetected (17). Among more than 800 β-lactamases identified in Gram-negative bacteria, at least 120 were detected in P. aeruginosa, among which, AmpC, ESBLs, and MBLs are clinically significant (8). The Current CLSI guidelines do not describe a method for detection of AmpC β‑lactamases. AmpC disc test was originally introduced to detect plasmid-mediated AmpC β-lactamases (18). However, Black et al. reported the detection of chromosomally mediated inducible AmpC β-lactamases in a number of bacteria including P. aeruginosa, by the AmpC disc test (19). In the present study, using the AmpC disc test, AmpC production (68.6%) was higher among the P. aeruginosa isolates from burns compared to those of other reports (17.3-59.4%) (17, 20-23). On the other hand, only half of the isolates which were positive in AmpC disc test showed AmpC production by TDET. Basak et al. also showed that the AmpC disc test was superior to TDET in screening for AmpC production (20).
The current study results showed a significant association between the AmpC phenotype determined by the AmpC disc test and AmpC gene carriage (P < 0.05). The rate of AmpC gene carriage (60.8%) in the present study was similar to a report from Taiwan, where presentation of AmpC gene in P. aeruginosa was 59.6% (16). However, in a study conducted in Iran, Fazeli et al. reported 100% AmpC gene carriage among 72 pediatric P. aeruginosa isolates from various clinical specimens, but did not show the AmpC phenotype (24). Gene presence does not necessarily mean its expression and depends on environmental conditions. Therefore, the current study believes that it is important to associate the enzyme phenotype with the presence of resistance genes.
Detection of ESBLs in AmpC producing Gram-negative bacteria is often a problem. High level expression of AmpC can prevent recognition of ESBLs leading to false negative results (25, 26). ESBL production in our isolates (39.2%) was higher compared to the other reports from Iran (2.2-23.3%) and other countries (1.81-17%) (16, 27-31). Coproduction of AmpC and ESBL (3.9%) in the isolates of this study was similar to the findings of Upadhyay et al. (3.3%), but much lower than several other reports (24.5-26%) (17, 22, 23). Finally, similar to a report from Brazil, the isolates of the present study did not harbor SHV and CTX-M genes (32).
In the past 30 years, extensive use of carbapenems to treat AmpC and ESBL producing P. aeruginosa, has led to increased levels of bacterial resistance mostly due to production of MBLs (9, 33, 34). Coproduction of AmpC and MBL was detected in 25.5% of the isolates which was low compared to the other studies worldwide (45.5-46.6%) (22, 23). Alarmingly, 11.8% of the isolates of the current study produced AmpC along with ESBL and MBL. Detection of multiple β-lactamases in highly resistant bacteria could be useful for the selection of suitable antibiotic therapy and avoiding treatment failure as well as reducing mortality rates in hospitalized patients.
In conclusion, the present study showed a high prevalence of AmpC production among P. aeruginosa isolates from burn wounds (68.6%). More importantly, multiple β-lactamase production was observed in 60% of the AmpC producers, the majority of which also produced MBL (25.5%). Co-production of AmpC with ESBL occurred in 3.9% of the isolates and 11.9% produced the three β-lactamases.