1. Background
Hepatitis C virus (HCV) infection is a major health problem that affects almost 3% of the world's population with a morbidity and mortality rates. Hepatitis C virus is a member of the Hepacivirus genus of the Flaviviridae family and the viral products (core, E1, E2, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) are processed from a 3000-amino acid (aa) polyprotein expressed from a single open reading frame (1). The E1 and E2 are enveloped proteins, which can elicit neutralizing antibodies against HCV infection in the host and Core, E1 and E2 proteins are the major vaccine candidates and Enzyme-Linked Immunosorbent Assays (ELISA) is one of routine tests in clinical laboratories and different studies to detect the rate of antibody in sera against HCV infection (2, 3). The combination of pegylated interferon α and ribavirin is a useful treatment depending on the viral or host factors but needs a prolonged therapy with different side effects (4).
Virus-like particles are self-assembled in the absence of DNA or RNA or genetic materials (2, 5). It has been shown that HCV antigens produced in Pichia pastoris induce strong immune responses in animals (6-10). In this study, the HCV VLPs obtained from P. pastoris hoping the rCoreE1E2 can induce neutralizing antibodies. Post-translational modifications such as proteolytic processing, folding, disulfide bond formation and glycosylation can be done by P. pastoris (11). This system is also faster, easier, and less expensive than expression systems derived from higher eukaryotes, such as insect and mammalian tissue cultures and usually gives higher expression levels (12-15). The expression systems can raise error rates in translation due to codon bias, that is, a preferential use of codons for the same amino acid (16, 17). As codon bias differs between the host organism and the organism from which the gene was extracted, substituting synonymous codons can improve translational fidelity and in this study codon optimization has been used for increasing the protein expression efficacy (18).
2. Objectives
In this study, the P. pastoris expression system was used to express a recombinant HCV CoreE1E2 protein, which consists of Core (269 nt-841nt) E1 (842 nt-1417nt) and E2 (1418 nt-2506nt).
3. Materials and Methods
3.1. Construction of Recombinant Expression Plasmid
The Core (269 nt-841nt) E1 (842 nt-1417nt) and E2 (1418 nt-2506nt) were amplified by Polymerase Chain Reaction (PCR) (94°C 5 minutes 1cycle, 94°C 30 seconds- 58°C 30 seconds- 72°C 100 seconds 30 cycles, 72°C 10 minutes 1 cycle ) from infected Iranian patient’s blood by HCV (genotype 1a) and primers were designed by Gene Runner software for Core forward [5- TT G A A T T C GTC A T G A G C A C G A A T C C T A A A C C T C -3], and E2 reverse [ 5- TT G C G G C C G C C G C C T C C G C T T G G G A T A T -3]. The amplified product was ligated into a pMD18-T vector (Takara, Japan) and cloned into the pPICZαA Vector (Invitrogen, Carlsbad, CA, USA) to produce pPICZαA-CoreE1E2. The clones in E. coli TOP 10 were obtained by transformation with CaCl2 and selected on in low salt LB medium with Zeocin™ (Invitrogen, USA) (11). Recombinants were confirmed by PCR, restriction enzyme digestion and sequencing. Then, the recombinant plasmids were linearized and electro-transformed into the competent P. pastoris cells performing as described by the instruction manual of P. pastoris expression kit (Invitrogen, USA).
3.2. Expression of rCoreE1E2 in Pichia pastoris
Briefly, pPICZαA-CoreE1E2 was linearized by Pme I and electroporated into P. pastoris strain GS115; GS115 (His −, Mut +) transformants were selected on Minimal Dextrose (MD) medium plate and confirmed on Minimal Methanol (MM) medium plate. Multiple inserted recombinants were isolated on Yeast Extract Peptone Dextrose (YEPD) medium plate containing Zeocin™ (Invitrogen, USA) at final concentration of 2.0 mg/mL. The P. pastorisGS115 strain was also transformed with the empty vectors pPICZαA for negative control tests. Clones were detected by colony PCR using the conditions and primers were provided in the EasySelect™ Pichia pastoris expression kit.
The control strains of the intracellular (GS115/β-galactosidase) and extracellular (GS115/albumin) expression provided by the EasySelect™ P. pastoris expression kit (Invitrogen, USA). After choosing the most resistant colon, a single colony of multiple inserted His −, Mut + GS115 recombinants was inoculated into 25 mL buffered glycerol complex medium (BMGY; 1% yeast extract, 2% peptone, 100 mM potassium phosphate (pH 6.0), 1.34% yeast nitrogen base (YNB), 4 × 10 - 5% biotin, 1% glycerol) and cultured at 250 rpm and 30°C until the culture medium reached an OD600 of 2 - 6. The cells were harvested by centrifuging and re-suspended in buffered methanol complex medium (BMMY; 1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% YNB, 4 × 10 - 5% biotin, and 0.5% methanol) to an OD600 of 1.0 and cultured in a 250 mL flask at 250 rpm under 30°C. Inductive expression was carried out with the addition of methanol (0.5%, v/v) per 24 hours at 30°C lasted for 3 days (1, 11).
The samples were analyzed by the Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Western blotting. For extraction and purification of rCoreE1E2, the cells were disrupted by glass beads in a Tris/EDTA/NaCl (TEN) buffer and then centrifuged. The deposit was re-suspended in a TEN buffer with 8M urea and centrifuged again. The supernatant was dialyzed against 5 mM Tris–HCl (pH 9.3) overnight at 4°C and then clarified by centrifugation. The supernatant was first loaded to a Q-Sepharose Fast Flow column with 20 mM Tris–HCl (pH 9.3). After washing with 0.05 M NaCl and 20 mM Tris-HCl (pH 9.3), the rCoreE1E2 was eluted with 0.5 M NaCl and 20 mM Tris–HCl (pH 9.3).Then the eluted was loaded to a Phenyl Sepharose Fast Flow column with 20 mM Tris–HCl (pH 9.3) and 0.1 M NaCl. The rCoreE1E2 was eluted with 20 mM Tris–HCl (pH 9.3). The rCoreE1E2 was further purified with Sephadex G150 column for removing low molecular weight fractions. The purified rCoreE1E2 was dialyzed against 5 mM Tris–HCl (pH 9.3) and sterilized with 0.45 m filter, and then stored at 4°C.
3.3. Protein Purification
Briefly, the supernatant was loaded onto Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, Germany) in the presence of 0.5% Tween 80 and 40 mM imidazole. The column was washed extensively, and the protein was eluted with 1 M imidazole. The rCoreE1E2 was purified from the insoluble fraction by using 0.5% N-lauryl sarcosine (Sarkosyl). The protein was purified over Ni-NTA super flow and the Sarkosyl was replaced by 0.1% Tween 80. The protein was eluted in the presence of 200 mM imidazole and 0.1% Tween 80 in phosphate buffer (11).
3.4. SDS-PAGE and Western Blotting
Supernatant of culture harvested and the cells were harvested and washed twice in TEN buffer. Cell disruption was performed by vortexing with glass beads in TEN. The lysate was clarified by centrifugation and the supernatant and pellet used. The rCoreE1E2 was mixed with sample buffer and boiled for 10 minutes in the presence of dithiothreitol (DTT), and around 200 ng was loaded on a 4 - 12% Bis-Tris NuPAGE gel (Invitrogen, USA). The primary antibodies against Core, E1 E2 were (Santa Cruse, USA); the secondary antibody was polyclonal rabbit anti-mouse horse radish peroxidase (HRP) labeled. Staining was done and the molecular weight marker was a mix of proteins from Sigma (USA): bovine serum albumin (A-7517), ovalbumin (A-7642), carbonic anhydrase (C-2273), beta lactoglobulin (L-4756) and alpha-lactalbumin (L-6385) or the Precision and Precision plus Protein Standards (Bio-Rad, USA).
3.5. Endoglycosidase Digestion Assay
Purified rCoreE1E2 was digested with N-glycosidase F (PNGase F) and the digested proteins were treated according to the manufacturer’s instructions (New England Biolabs,UK) and then analyzed by Western blotting with mAb fore Core E1 E2.
3.6. Animals, Immunization and ELISA
Approximately rCoreE1E2 (300 µg) diluted in 2 mL sterilized 0.9% NaCl was used for immunization. A New Zealand rabbits was subcutaneously immunized in multiple sites on the back with 0 g (negative control) or 300 µg purified rCoreE1E2, respectively. Booster injections were given with the same doses at 2, 3, 4, 5 and 13 weeks later. An indirect ELISA was used to measure anti-rCoreE1E2 antibodies in rabbit’s serum. In brief, a recombinant protein (10 mg/mL) diluted in 50 mM carbonate buffer (pH 9.6) was used to coat microtiter plates, overnight at 4°C. After blocking with 2% (w/v) skim milk powder (Oxoid Ltd,) in PBS with 0.05% (v/v), Tween 20 (Sigma, USA) (PBST), pH 7.2 for 1 hour at room temperature. The serum samples were added in duplicate, either 1/20 in dilution buffer (PBST), containing 1% (w/v) skim milk powder, to test seroconversion or in a double serial starting at 1/50 dilution for titration. They were incubated at 37°C for 1 hour. A HRP-labeled antirabbit IgG (Sigma, USA) was added to 1/10,000 in the dilution buffer. After 1 hour of incubation at room temperature (RT) and washing, tetramethylbenzidine substrate (Sigma, USA) reactions were stopped with 50 mL of 2.5 M H2SO4. Absorbance was read at 450 nm in a SensIdent Scan (Merck, Germany). The cut-off value used to consider a positive antibody response was established as twice the mean of absorbance values of the preimmune sera (19).
3.7. Enzyme-Linked Immunosorbent Assays With Human Sera
HCV rCoreE1E2 particles were coated at 1 g/mL in carbonate buffer in microtiter plates and were incubated. The plates were washed and incubated with blocking buffer (0.5% casein in PBS). Sera of chronically HCV-infected people or sera of healthy persons were added in a dilution of 1/20. The plates were washed and incubated with horse radish peroxidase labelled rabbit anti-human immunoglobulins. The tetramethylbenzidine substrate (Sigma, USA) reaction was stopped by addition of 2 N H2SO4 and the absorbance was measured at 450 nm. The cut-off value to consider a positive antibody response was established as twice the mean OD 450 nm of the negative control sera (19-21).
3.8. Transcriptional Analysis of the CoreE1E2 Gene
Total RNA was extracted and cDNA was synthesized using the IMPROM-II™ Reverse Transcription System (Promega, USA). The presence of a heterologous gene mRNA in Pichia was detected by the Real Time-Polymerase Chain Reaction (RT-PCR) and Real-time PCR with Forward primer 5′- TTGGGACATGATGATGAATTGG -3′ and Reverse primer 5′- TGCCTGTGGGATTCTAAGC -3′ and probe 5′- ACAGCCGCATTGGTTGTCGCC -3′ that anneal in the internal sequence of the CoreE1E2 genes (21, 22).
3.9. Codon Optimization
The codon-optimized gene was designed based on the protein sequence of CoreE1E2 according to the codon bias of P. pastoris (http://www.kazusa.or.jp/codon). Codon optimization was performed by using the GenScript program. The entire CoreE1E2 gene with Xho I and Not I restriction sites at each end was designed and was in frame with α-factor of a pPICZαA vector. The designed rCoreE1E2 was synthesized by (GenScript, USA). Our supernatant collected and recombinant CoreE1E2 was purified and fixed in glutaraldehyde and negatively stained with the uranyl acetate prior to analysis by the transmission electron microscopy.
4. Results
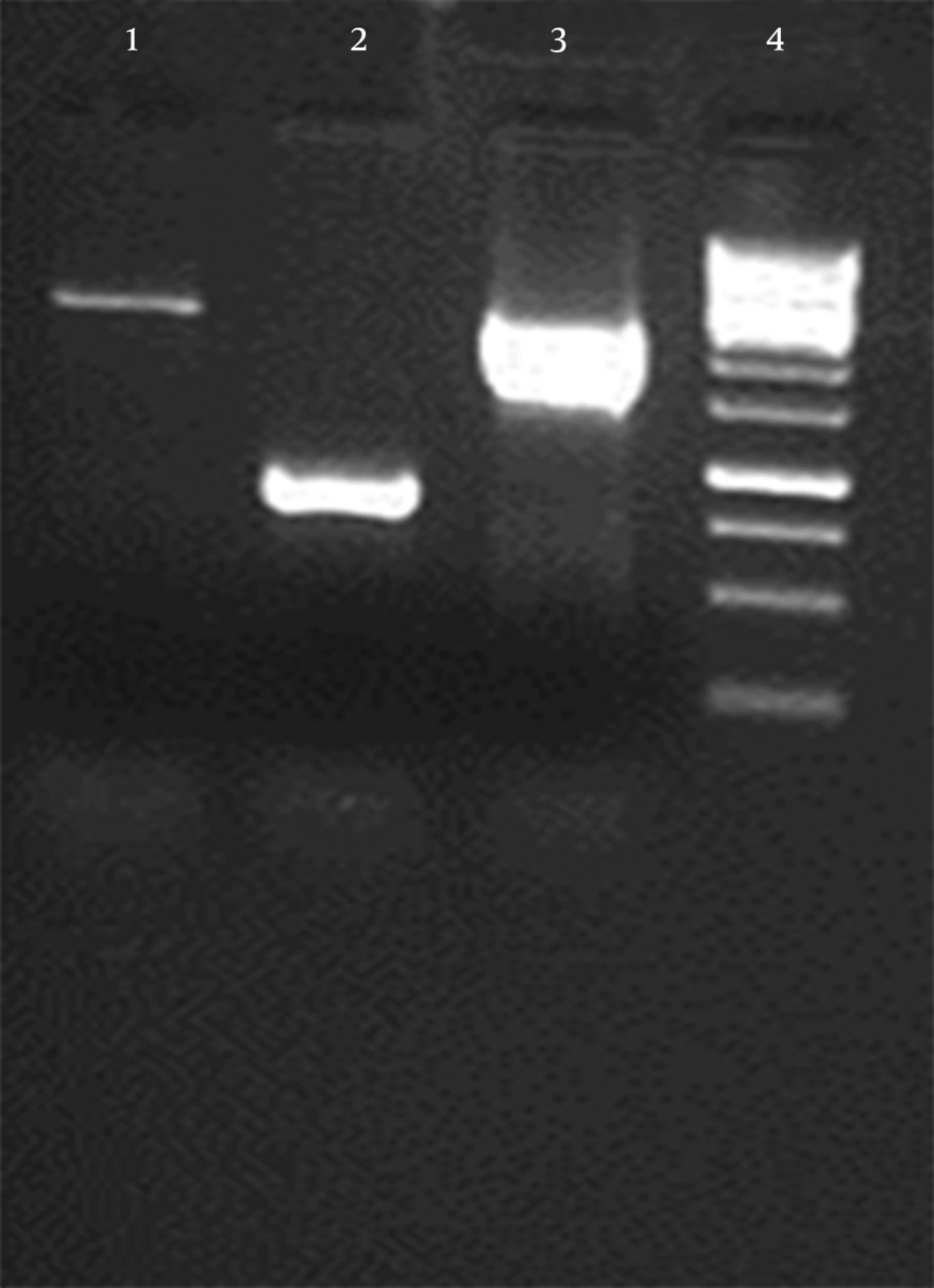
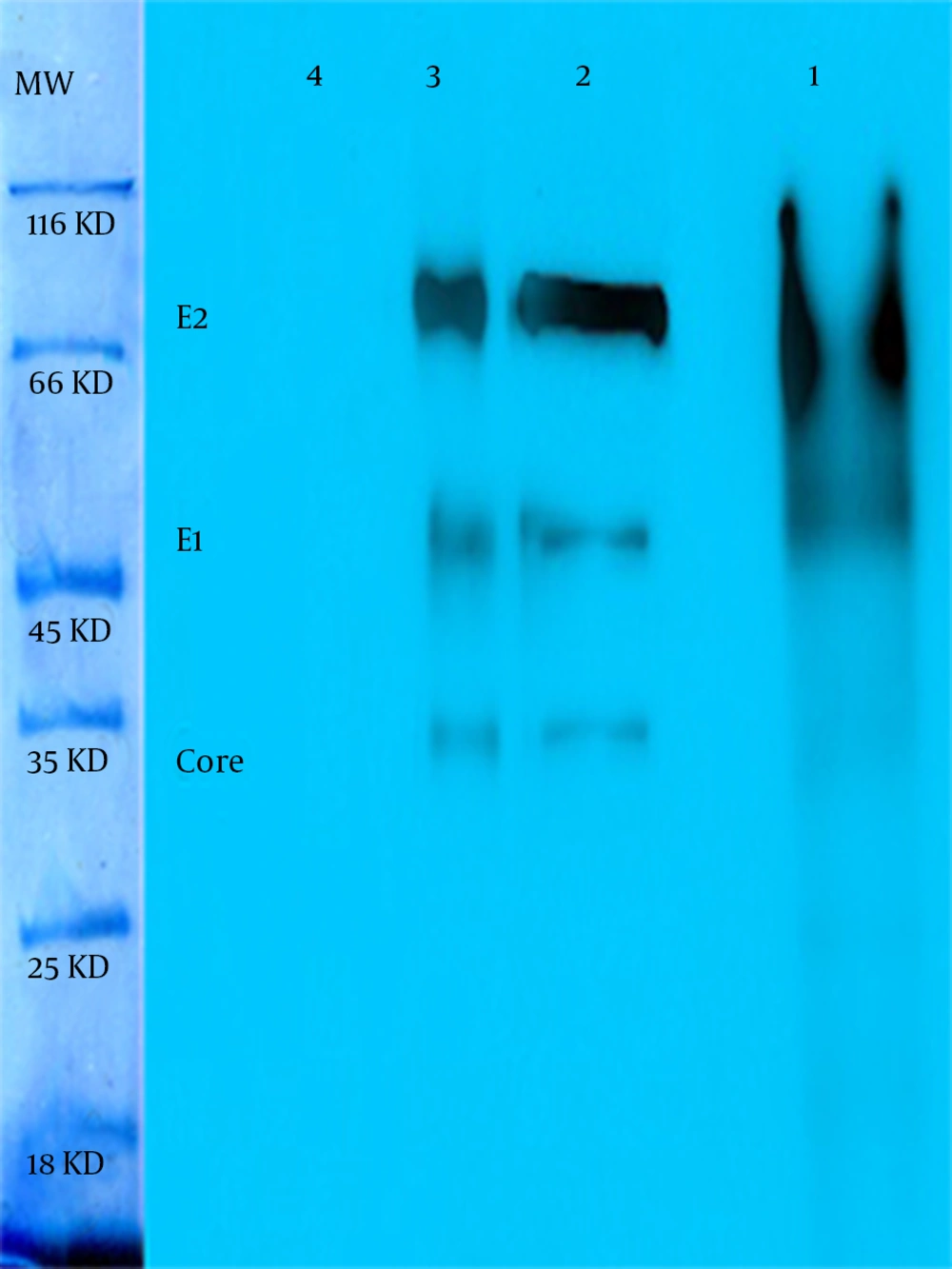
The continuous coding regions of Core (269 nt-841nt) E1 (842 nt-1417nt) and E2 (1418 nt-2506nt) were amplified by PCR from infected blood by HCV (genotype 1a) and the product (2237 bp) was cloned in a vector. The colony PCR was done to verify the insertion of target gene in vector (Figure 1). After that target gene and pPICZαA digested by XhoI and NotI enzymes and ligated into recombinant expression vector The pPICZαA-CoreE1E2 was linearized and electroporated into P. pastoris strain GS115; GS115 and different colonies in different concentrations of Zeocin were evaluated and the most resistant colons, which tolerated 800 and 1600 mic/mL were chosen for a huge expression phase. The rCoreE1E2 protein was purified and proteins were studied by SDS-PAGE and Western blotting (Figure 2 and 3). Core protein with 20 kDa and E1 protein with 40 kDa and E2 with 60 kDa were shown. The codon optimization was designed based on the protein sequence of CoreE1E2 according to the codon bias of P. pastoris (http://www.kazusa.or.jp/codon).
The virus like particle is shown in our supernatant with electron microscopy (EM) negative staining image with 70 nm particles in our sample. The value of Codon Adaptation Index (CAI) of 1.0 is considered to be ideal and perfect while a CAI of > 0.80 is rated as good for expression in the desired P. pastoris. The ideal percentage range of GC content is between 30 to 70%. The percentage distribution of Codon Frequency Distribution (CFD) of 100 is set for the codon with the highest usage frequency for a given amino acid in P. pastoris. Codons with values lower than 30 are likely to hamper the expression efficiency and as it shows after codon optimization the value of Frequency of Optimal Codons (FOP) in our gene is acceptable and all cordon’s distributions are upper than 50%. Interestingly, 60 percent of Codon Frequency Distribution is 91 - 100%, 5 percent is 81 - 90%, 8 percent is 71 - 80%, 6 percent is 61 - 70%, 14 percent is 51 - 60% and 3 percent is 41 - 50% and no distribution lower than 40% was detected.
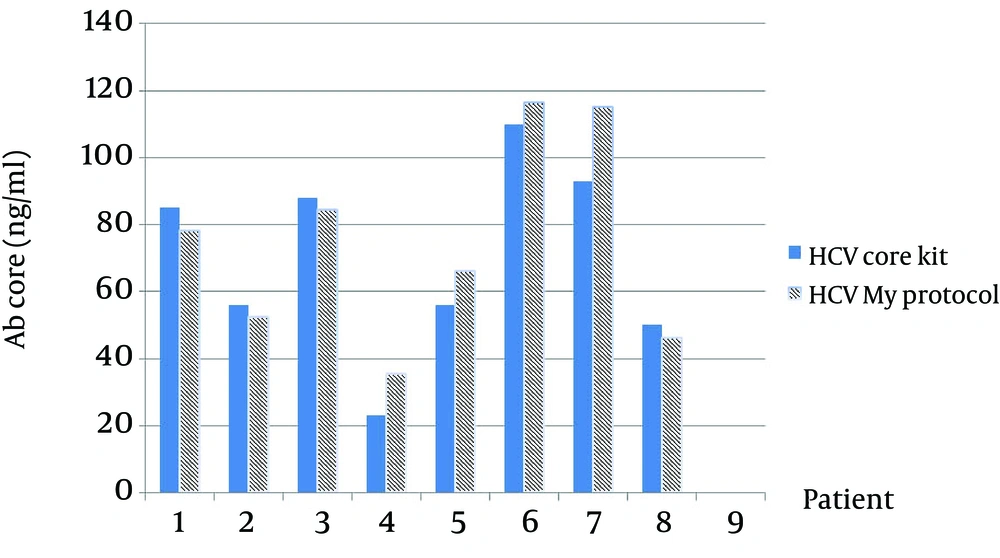
As in Figure 3 is clearly shown, the thickness of bands in an optimized sample comparing with a not-optimized sample shows that most codons are expressed better in P. pastoris. The presence of target mRNA in P. pastoris was detected by RT-PCR and Real-time PCR to detect Core-E1-E2 and housekeeping genes (data not shown). To digest carbohydrate residues from a recombinant Core-E1-E2, PNGase F was used to remove unnecessary N-glycan bands (Figure 4). As it is clear, before digestion the presence of smear in lane 1 is present but after digestion in lane 2 and also in positive control, all bands are sharp and Core with 20 kDa and E1 with 40 kDa and E2 with 60 kDa are illustrated. For evaluation rCoreE1E2 particles against patient sera, we compared our homemade kit with international QuickTiter™ HCV Core Antigen ELISA Kit (Catalog Numbers VPK-151) against the sera of some patients (Figure 5). From those patients, 8 samples were shown and interestingly in case number 4, 5, 6, 7 the result of our kit is better than a standard kit.
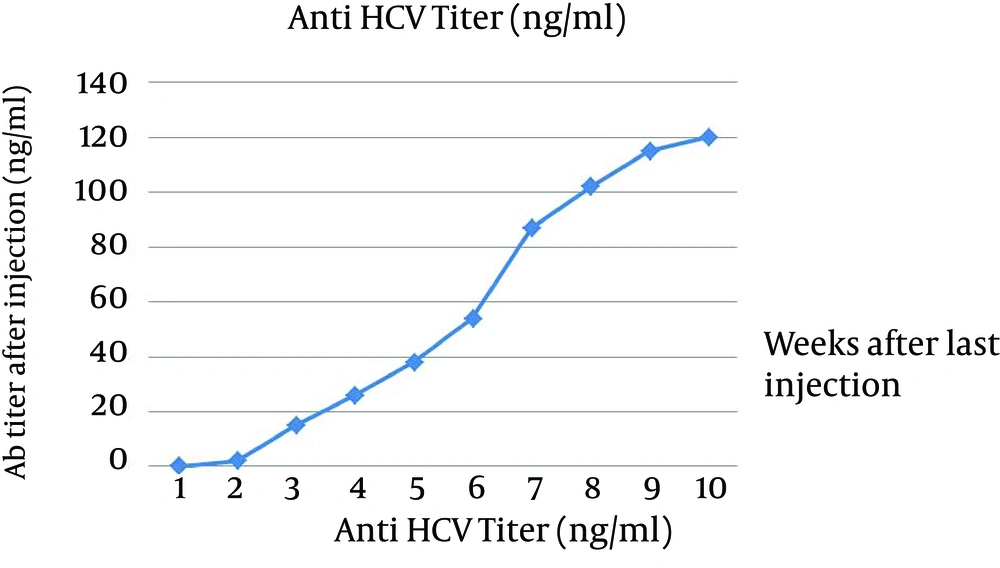
Our data were repeated 3 times and all results were the same. Number 7 has the highest titer of antibody and in our kit it is 118 ng/mL but in QuickTiter™ kit it is 90 ng/mL and in case 4 the lowest titer was detected and antibody level by our kit shows 38 ng/mL and QuickTiter™ kit shows 22 ng/mL. For in vivo immunization, the New Zealand rabbit was subcutaneously immunized in multiple sites and after a last booster injection the titer of antibody was checked. The first serum was collected before immunization and no anti HCV antibody was detected by ELISA and this sample was kept as zero in the data chart. After the last injection, every week sample showed the raise of antibody; for example in week 5 the rate of antibody is 49 ng/mL in week 8 is 100 ng/mL and surprisingly in the week of 10 the antibody titer is 120 ng/mL (Figure 6).
The blue bar is the result of antibody of patients antigen ELISA Kit QuickTiter™ HCV core and another bar is antibody titers of patients with our ELISA kit; In this chart 8 samples are shown and in number 4, 5, 6, 7 the result of homemade kit is better than QuickTiter™ kit; data were repeated 3 times and results were the same.
5. Discussion
One of the aims of HCV research is to develop an effective vaccine to produce acceptable immunity in human sera against HCV glycoproteins (2, 6). In our study, the Iranian sample of HCV virus (genotype 1a) diagnosed and used as a temple for amplification. Although some research showed that it was difficult to express HCV envelope proteins by yeast P. pastoris (11), we could express HCV rCoreE1E2 by P. pastoris expression system. The expression vector, which was used in our study is a pPICZαA vector and this vector has α-factor to help us to express and secret recombinant protein in high volume. Therefore, Western blotting using anti-Core/E1/E2 mAbs demonstrated a different bands for Core 20 kDa, E1 40 kDa, E 260 kDa in yeast-expressed system. Using a eukaryotic expression system for HCV envelope glycoproteins can help us to make native shape and function for our recombinant proteins.
Pichia pastoris can synthesize and process rCoreE1E2 carrying glycans, which could be digested by PNGase F and this glycozilation is similar to some of original HCV envelope glycoproteins. The truth is that glycans of rCoreE1E2 should be different to native CoreE1E2 because of the difference between yeast and mammalian cells. The PNGase F removes all three types of amino-linked glycans, high mannose-type glycans, hybrid - type glycans, and complex - type glycans. In P. pastoris cells, the N-glycosylation pathway is similar to the pathway in human cells except that P. pastoris cells have just high mannose structures. Enzymatic deglycosylation with PNGase F resulted that the glycosylated smeared band was disappeared and sharp band with less molecular weight, which corresponds to nonglycosylated proteins was remained, indicating that the multiple bands arise by different degrees of N-glycosylation. The yeast expressed rCoreE1E2 has all potential N-glycosylation sites occupied (11, 19). We reported that codon optimization leads to increase the expression of recombinant Core-E1-E2 in P. pastoris. We designed the Core-E1-E2 gene by choosing the most preferred codons, while avoiding the formation of stable secondary structures and our data was similar with other optimization studies, which show an increase of efficacy by codon optimization (16-18).
The translational hypothesis related to translation initiation and elongation rates has been well- accepted for explaining the codon usage bias in eukaryotes. Although the mRNA levels were similar between the native and the optimized constructs, suggesting that the increased expression is attributable to the enhancement of posttranscriptional processing (data not shown). As the genes were placed after the α-factor secretion peptide, we expect that the increased expression by codon optimization should be mainly due to the enhanced translation elongation instead of translation initiation. It seems that other factors like protein folding within the endoplasmic reticulum and secretion signal processing may be important in secretion ability. Moreover, in our study the native gene employs tandem rare codons that can reduce the efficiency of translation or even disengage the translational machinery. We changed the codon usage bias in P. pastoris by optimizing the CAI to 0.85. GC content and unfavorable peaks have been optimized to prolong the half-life of the mRNA. The Stem-Loop structures, which impact ribosomal binding and stability of mRNA, were broken. In addition, our optimization process has screened and successfully modified those negative cis-acting sites.
In other past researches, different parts of HCV glycoproteins were used for immunization in mice, goat, sheep and raising antibody detected (6, 9, 19, 23). In this study, the strategy of inducing broadly neutralizing antibodies is probably successful to produce anti HCV glycoprotein antibody as it succeeds in rabbit and our rCoreE1E2 can induce high humeral immune response and it can be one step forward for evaluation of HCV vaccine for in vivo research. The immune reactivity of rCoreE1E2 particles was tested by the international ELISA Kit QuickTiter™ HCV and homemade ELISA kit, using sera from chronically HCV-infected persons. Indicating the epitopes presented by our particle’s conformation is very analogous with the original HCV particle. The evaluation of human sera, showing anti-HCV positive sera against rCoreE1E2 proteins demonstrated that anti-HCV positive sera recognized our recombinant peptide by ELISA and even in some cases our results were better than the international kit. Moreover our data shows that human sera is anti-recombinant protein and can neutralize our protein in ELISA system which has better result than other researches and all immunogenic sites are in our recombinant protein, which has not been in other studies (20, 24). The virus like particles have a modal diameter centered about 70 nm and were shown with negative staining by Electron microscopy and also because CoreE1E2 assembled together, the size of particle increased which is similar to other studies base on HCV particles in vivo and in vitro (7, 8, 13).
In conclusion, the expression of the HCV structural proteins in P. pastoris would be useful for studying the mechanisms of HCV processing, morphogenesis, immunity and assembly. Natural HCV structural proteins are not useful for developing vaccines or specific anti-sera because the virus concentrations in the infectious materials are very low. Therefore, recombinant HCV structural proteins are useful as immunogens. For the development of preventive vaccines and therapeutic treatments against HCV, the rCoreE1E2 protein might be a crucial element and the results obtained in this work may therefore contribute to this effort. These recombinant proteins may be useful targets for HCV vaccine candidates. Moreover, P. pastoris yeast expression system is an efficient eukaryotic expression system and we believe that the P. pastoris yeast-expressed rCoreE1E2 is a promising HCV vaccine candidate for industrial purpose.