1. Background
Over the past decades, the incidence of mucormycosis (zygomycosis) infections has increased. Mucormycosis is a fungal infection caused by Mucorales. The members of this order with variable antifungal susceptibility patterns include Mucor, Absidia, Rhizopus, Rhizomucor, and Lichtheimia. This order is environmental fungi with rapidly progressive and life-threatening infections in immunocompromised patients with underlying diseases like diabetes mellitus, neutropenia, bone marrow transplants, hematological malignancies, and advanced AIDS (1). Interestingly, there are reports of patients with this infection with no immune compromising conditions (2, 3).
Infection could be caused by inhaling aerosolized Mucor spores from the soil and the environment. The most clinical features of infections are rhinocerebral, pulmonary, and disseminated disease. Classic clinical features are fever, pain, headache, facial swelling, nasal congestion, ocular involvement with visual loss, and limitation of eye movement. In pulmonary or disseminated infections, the signs and symptoms are similar to those of other fungal or bacterial infections (1, 4).
The mortality risk of these infections varies and depends on different characteristics, such as patient population and site of infections (1, 4). In critically ill adult patients admitted to the intensive care units during 2007 to 2015, mucormycosis was responsible for 25.8% of fungal infections (5). The mortality rate of infections was extremely high and was reported as 38% to 100% in immunocompromised patients, such as those with hematologic malignancy and hematopoietic stem cell transplantation, and transplant recipients (6-10). Roden et al. demonstrated 76% mortality in patients with pulmonary infections despite aggressive surgical and antifungal therapy (11). Currently, mucormycosis is mostly established by culture yet the sensitivity of this method is low (8-11). As there are reports about resistant species of this genus to the suggested treatments, identification of causative agents could play an important role in the management of infected patients.
2. Objectives
The aim of the present study was the identification of etiologic agents of respiratory tract mucormycosis based on sequencing methods using 18S rDNA, for the best management of infected patients.
3. Methods
3.1. Ethics Statement
The ethics committee of the clinical microbiology research center, Shiraz University of Medical Sciences, approved this study (EC-958784), which was carried out in accordance with the Declaration of Helsinki.
3.2. Study Patients
A total number of 163 patients consisting of 70 (43%) females and 93 (57%) males were included in the present study. Patients were suspected of invasive fungal diseases based on clinical and radiological features. Sinus tissue, bronchoalveolar lavage (BAL), and blood samples were collected during 2012 to 2015. This sampling was part of the diagnosis processing. All the samples were transferred to the Mycology division of Professor Alborzi clinical microbiology research center, Namazee hospital, Shiraz University of Medical Sciences, Shiraz, Iran. Demographic data from all the patients such as age, gender, background disease, duration of symptoms and previous medical history, and pathology results were obtained from the patient’s medical files.
3.3. Conventional Methods
Sinus tissue and BAL samples were examined by microscopic examination using 10% potassium hydroxide (KOH) for the detection of hyphae. Also, they were cultured on Sabouraud dextrose agar (Merck, Darmstadt, Germany) and incubated for 14 days at 30°C. Blood samples were cultured on BACTEC medium (Becton-Dickinson, Sparks, MD, USA).
3.4. Molecular Method
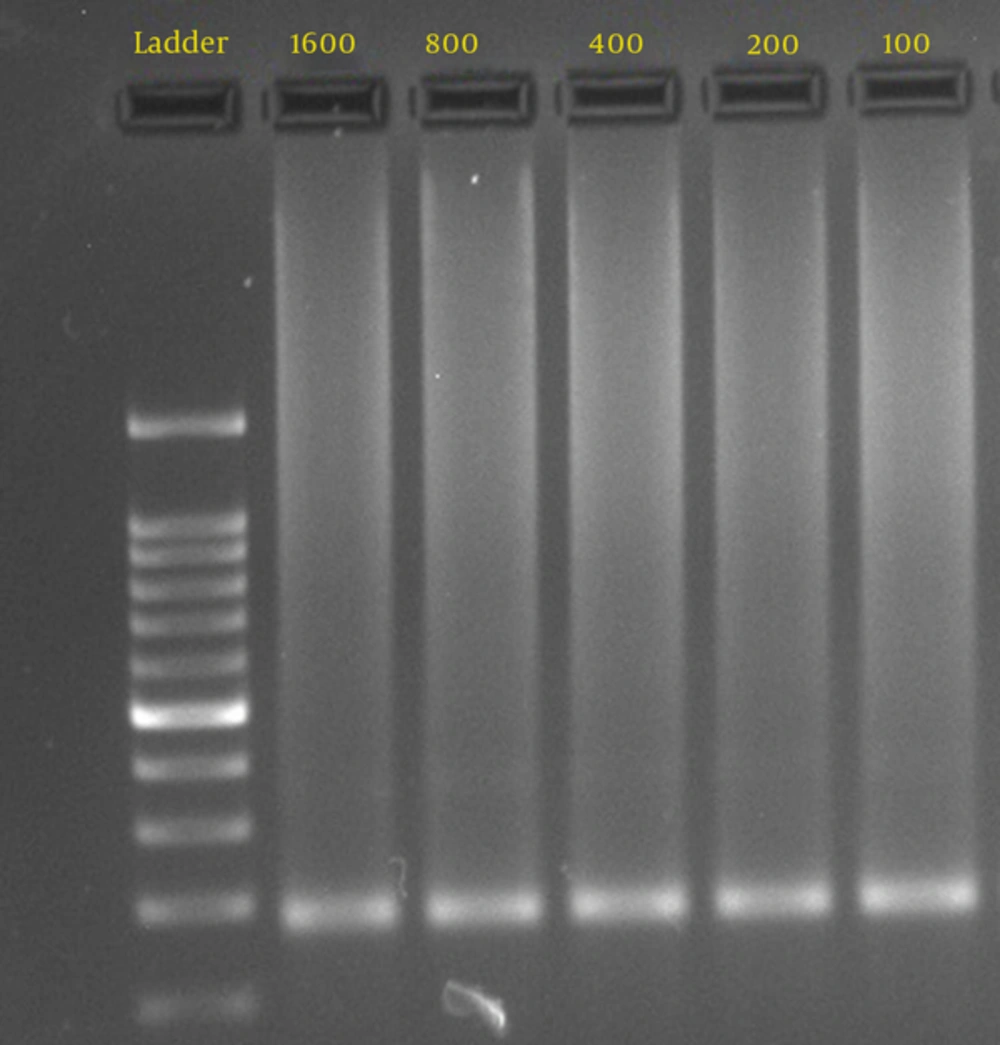
To determine the sensitivity of the PCR assay, DNA was extracted from the culture of Mucor species isolated from the patients (Figure 1). Semi-nested PCR for diagnosis of mucormycosis was performed on all samples (blood, tissue, and BAL) and Mucorales isolated from culture media. For DNA extraction and tissue lysing, 100 µL of distilled water, 100 µL of lysis buffer, 20 µL of proteinase K and 20 µL of carrier RNA were added to the tube containing tissue samples, incubated overnight at 56°C, and washed with normal saline. The BAL samples were centrifuged at 3000g for 10 minutes. DNA was extracted using a DNA extraction kit (Invisorb® Spin DNA Extraction Kit, Berlin, Germany), as recommended by the manufacturer. To avoid contamination, all lab procedures were handled under sterile conditions in biological safety cabinet class 1. Semi-nested PCR was used according to Rickert et al. (7). Two sets of primers: ZM1 (5́-ATT ACC ATG AGC AAA TCA GA-3́) and ZM2 (5́-TCC GTC AAT TCC TTT AAG TTT C-3́) for the first round of PCR and ZM1 and ZM3 (5́- CAA TCC AAT TTC ACC TCT AG-3́) for the second round were used.
The first reaction mixture consisted of 10 µL of extracted DNA, 10x PCR buffer (CinnaGen, Iran), 1 µM specific primers (Bioneer, South Korea), 1.5 units Taq DNA polymerase (CinnaGen, Iran), 100 µM dNTPs mix (Zist Asia, Iran), and 2.5 mM MgCl2 (CinnaGen, Iran). The total volume of master mix was 50 µL. The second reaction mixture was identical, except that instead of the DNA extract, 5 μL of the product from the first round was used. All PCRs were run in a thermocycler (Mastercycler, Eppendorf, Hamburg, Germany), under the following conditions: denaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 30 second, annealing for 30 seconds at specific temperature (55°C), and extension at 72°C for 8 minutes, which amplified a product of 176 to 177 bp. The PCR products were analyzed by electrophoresis in 2% agarose gels, and visualized with the Geldoc system (Gel logic 200, Kodak. USA). Products of positive PCR were sequenced and viewed with Chromas version 2.24 software (Technelysium Pty., Ltd). Then, the sequences were used for BLAST search in the Gene bank database (National Center for Biotechnology Information, National Institutes of Health, and Washington DC, USA).
4. Results
The median age of the patients was 39 years (2 to 75 years). The most common risk factor in patients was having diabetes mellitus, followed by bone marrow transplant, acute myelocytic leukemia, acute lymphocytic leukemia, and other hematopoietic disorders. The limitation of detected DNA concentration by nested PCR was 100 fg (Figure 1). Direct microscopic examinations of tissue invasion, culture, and semi-nested PCR were positive in 11.7% (19/163), 6.7% (11/163), and 10.4% (17/163) of patients, respectively (Table 1). In the present study, the result of the pathologic examination was considered as the gold standard. In patients with a positive pathologic examination, culture and semi-nested PCR positive results were observed in 11/19 (57.8%) and 17/19 (89.5%) patients, respectively. None of the blood cultures were positive for Mucorales. According to direct smear and culture results, all the identified isolates were in the Mucoraceae family and identification by genus was difficult.
| No | Age/Sex | Direct Smear | Culture | Semi-Nested PCR | Sequencing of PCR Products |
|---|---|---|---|---|---|
| 1 | 35/F | Positive | Positive | Positive | R. oryzae |
| 2 | 19/F | Positive | Negative | Positive | R. oryzae |
| 3 | 28/M | Positive | Positive | Positive | R. microsporus |
| 4 | 56/F | Positive | Positive | Positive | R. oryzae |
| 5 | 65/F | Positive | Negative | Negative | - |
| 6 | 25/M | Positive | Negative | Positive | R. oryzae |
| 7 | 48/M | Positive | Positive | Positive | R. microsporus |
| 8 | 32/M | Positive | Negative | Positive | R. oryzae |
| 9 | 40/F | Positive | Positive | Positive | R. microsporus |
| 10 | 5/F | Positive | Positive | Positive | R. oryzae |
| 11 | 16/M | Positive | Negative | Negative | - |
| 12 | 55/F | Positive | Negative | Positive | L. corymbifera |
| 13 | 71/F | Positive | Positive | Positive | R. oryzae |
| 14 | 66/M | Positive | Positive | Positive | R. microsporus |
| 15 | 45/F | Positive | Negative | Positive | L. corymbifera |
| 16 | 10/M | Positive | Positive | Positive | R. oryzae |
| 17 | 36/F | Positive | Positive | Positive | R. microsporus |
| 18 | 63/M | Positive | Negative | Positive | R. oryzae |
| 19 | 74/M | Positive | Positive | Positive | R. oryzae |
a Rhizopus: R, Lichtheimia: L
The results from molecular sequencing methods correlated well with automated microbiological identification systems for common clinical isolates. The results of DNA sequencing were compared with data in Gene Bank. The etiologic agents were Rhizopus oryzae (10 cases), R. microsporus (5 cases), and new species (2 cases). The sequencing of these new genes demonstrated 98% identity with Lichtheimia (Absidia) corymbifera genus with accession numbers: JQ775460.1. This new sequence (645 bp) was published in Gene Bank with accession number KJ690940 (Uncultured Mucor clone IR-B-Ch 18S ribosomal RNA gene, partial sequence) and European Nucleotide Archive of EMBL-EBI with accession number LT555571 (12, 13). The lineage hierarchy of the organism lies under Eukaryota, Fungi incertae sedis, Mucoromycotina, Mucorales, Mucorineae, and Mucoraceae, respectively. Table 2 shows the phylogeny reconstruction analysis based on lineage report of nucleotide blast for the new sequence. Unfortunately, we could not identify these fungi from culture media.
| Organism | Sample Gene |
|---|---|
| Eukaryota | |
| Fungi | |
| Mucorales | |
| Uncultured Mucor | KJ690940.1 |
| Lichtheimia corymbifera | JQ775460.1 |
| Lichtheimia hyalospora | JQ775448.1 |
| Lichtheimia sphaerocystis | JQ775447.1 |
| Lichtheimia ornata | JQ775432.1 |
| Lichtheimia ramosa | JQ775458.1 |
| Absidia sp. MJ3-3 | HM590653.1 |
| Absidia sp. MJ2 | HM590652.1 |
| Uncultured fungus | KP663784.1 |
5. Discussion
The incidence of mucormycosis has increased in the recent decades, yet its diagnosis remains difficult (14). The most common risk factor of the disease in this study and other studies was having diabetes mellitus (3, 15). Direct smear with KOH or pathology smear are rapid methods for the diagnosis of infection and in the present study, this method was considered as the gold standard test. Direct microscopic examinations in this study (histopathology and KOH) were positive in 19/163 cases (11.7%) and Rickerts et al.’s study reported that 5/21 (23.8%) immunocompromised patients had positive results by histopathological examination (7). Histopathological examination cannot identify the accurate etiologic agent; therefore, treatment is in the challenge.
The sensitivity of culture and identification of specific species of Mucorales in culture was reported to be variable, and in some positive cases documented by histopathology, culture may become negative. The sensitivity of culture for this genus was reported as 20% to 50% (7, 11). In the present study, 11/163 (6.7%) of patients had positive culture results. When the fungi were isolated from culture media, there was 21% discrepancy in the determination of Mucorale between morphological and sequence-based methods (16). It seems that the identification of species by sequencing could be important to the geographic distribution of mucormycosis. Incorrect identification of etiologic agents leads to wrong treatment of patients.
In the present study, the 18S rDNA of fungi belonging to the order Mucorales were used to diagnose the infection in clinical samples and species-specific by sequencing. The results of PCR demonstrated that 17/19 (89.5%) patients were documented by positive PCR. The sensitivity of PCR for diagnosis of mucormycosis was reported as 100% (5/5) in the study of Rickerts et al. (7). Polymerase chain reaction is of greater value in tissues than in other specimens like blood (17, 18) and endoscopic sinus surgery specimens (18). The different results are due to the type of molecular methods and number of specimen used in each study. The predominant etiologic agents are different in each region; in this study, Rhizopus species (88.2%) was the most prevalent and in the study of Samarei et al., among the identified species, the rate of Rhizopus species was 26.7%, and 40% (12/30) of samples were not identified (19). New strains in this study were with 98% identity with L. corymbifera. In Ziaee et al.’s study, 9.17% of 120 pure Mucorale cultures belonged to L. corymbifera (20). Absidia corymbifera is the pathogenic species of the genus Absidia reclassified as L. corymbifera (21, 22).
Lichtheimia species are the second cause of mucormycosis in Europe and third worldwide (23, 24). In Iran, there has been limited reports about this organism due to misdiagnosis. These findings are important in the treatment of patients with mucormycosis. Pulmonary Lichtheimia infections in solid organ transplant patients seems to be a higher risk for the development of disseminated infection (25). Also, Lichtheimia, which is associated with Farmer’s lung disease (a hyper sensitivity disorder) is observed among people in farming jobs and agriculture with frequent contact with contaminated material (26). It is the etiologic agent of some infections in immunocompromised humans and animals (pulmonary, central nervous system, rhinocerebral, or cutaneous) with a mortality rate of about 100% in HIV/AIDS patients (27, 28).
Schwartze et al. evaluated genomics analysis and secreted material of L. corymbifera and R. oryzae (the most prevalent isolated agents from patients), and reported many differentiations in genetic and the number of secreted proteases in these organisms (29). The members of Mucorales exhibit variable susceptibility patterns to antifungal agents and identification of the causative agent to the species level could be beneficial to the treatment of patients. For example, it was reported that “Rhizopus species were significantly less susceptible to itraconazole, posaconazole, terbinafine, and amphotericin B than Absidia species and less susceptible than Mucor species to amphotericin B” (28).
6. Conclusion
Sufficient information about etiologic agents in each region by the molecular method and DNA sequencing could be used for the best management and treatment of infected patients. For the diagnosis of infections, histopathology would be most efficient yet cannot identify the etiologic agents as species. Diagnosis of the new species may be considered as future antifungal therapy and management of the patients.