1. Background
Acanthamoeba is a prevalent genus of free-living amoebae, found in various environmental sources. Human infection with Acanthamoeba can result in brain, eye, skin, and lung diseases (1). Isolation of Acanthamoeba cysts from environmental sources, such as soil, dust, and stagnant water, shows that environmental sources play an important role in the transmission of amoebae to humans and other mammals (1, 2).
Acanthamoeba transmission is possible through the nostrils, skin lesions, and contact lenses. Immunocompromised and AIDS patients are susceptible to Acanthamoeba infections. These infections may cause acute diseases, such as granulomatous amoebic encephalitis (GAE), skin lesions, and pneumonia, which may lead to death (1). In addition, amoebic keratitis has been reported in contact lens wearers and may lead to blindness (1).
Molecular methods, especially polymerase chain reaction (PCR) on variant regions of 18s rRNA genes, are used for identification and taxonomy of Acanthamoeba species. According to the sequencing of 18s rRNA genomes, 20 Acanthamoeba genotypes (T1-T20) have been identified (1, 3). Different studies have shown that T4 is the most common genotype of Acanthamoeba spp. to cause amoebic keratitis and GAE (1).
T4, T6, and T2 genotypes of Acanthamoeba spp. have been abundantly reported in fresh and stagnant water, soil, and warm water of Iran (4, 5). According to previous studies, Acanthamoeba is present in 25.4% to 71.6% of water samples from East Azerbaijan and Ahwaz, Iran (6, 7). In addition, 26% of soil samples from Ahwaz and Tehran contain Acanthamoeba spp. (6, 8). Niyyati in another study from Iran showed that all 13 dust samples contained T4 genotypes, which can cause amoebic keratitis (9). The prevalence of Acanthamoeba spp. was 42.4% in water samples from Zabol, Iran (10) and 84.6% and 58.6% in water and soil samples from Pakistan, respectively (11). Also, in Brazil, 55% of stagnant water samples were infected with Acanthamoeba (12).
Because of specific weather conditions and dust storms in Kashan, people, especially immunocompromised patients, are at a greater risk of infection due to free-living amoeba and Acanthamoeba. Furthermore, mosques have many ablution pools, which may transmit amoebae, responsible for GAE and amoebic keratitis.
2. Objectives
Considering the role of Acanthamoeba spp. in GAE and amoebic keratitis (1) and transmission of this organism via water, soil, and dust containing amoebic cysts (1), we aimed to identify the genotypes of Acanthamoeba spp. in soil, hospital dust, and water samples from Kashan, Iran. To the best of our knowledge, no similar studies have been conducted regarding the prevalence and genotyping of Acanthamoeba in different environmental sources of Kashan, Central Iran.
3. Methods
3.1. Ethics Statement
The ethics committee of Kashan University of Medical Sciences approved this study (IR.KUMS.Rec.1394.126).
3.2. Study Setting
Kashan is a city located in the North of Dasht-e Kavir. It is a historical city with attractive sights in the Northwest of Isfahan Province, Central Iran. Kashan, with a population of nearly 400,000 people, is situated in 51 degrees 27 minutes east longitude and 33 degrees 59 minutes north latitude (altitude from the sea level, 950 m). The mean temperature is 19°C, which can reach as high as 50°C. The annual rainfall has been averaged as 137 mm, and the average humidity is 37%; stormy winds are common in Kashan (13).
3.3. Sample Collection
In this cross sectional study, a total of 122 samples were randomly collected from numerous environmental sources, including soil (n, 32), hospital dust (n, 40), and stagnant surface water (n, 50) from Kashan during February 2015 and December 2016. The samples were transferred to the Department of Parasitology of Kashan University of Medical Sciences.
The soil samples (12 g) were obtained from Kashan parks, while dust samples were collected from the immunodeficiency wards of Shahid Beheshti hospital (including the infectious diseases and chemotherapy wards), the Ear, Nose, and Throat (ENT) Center of Matini Hospital, and Hemodialysis Unit of Akhavan Hospital. The samples were investigated for the presence of free-living amoebae and Acanthamoeba strains. In addition, 300 mL of stagnant water from pools in parks was checked for the presence of these organisms.
3.4. Filtration and Cultivation of Free-Living Amoebae
The soil and hospital dust samples were dissolved in 100 and 20 mL of sterile distilled water, respectively, while the supernatants and stagnant water samples were filtered using a pumping machine (AEI, UK) with a 0.45-μm cellulose nitrate filter (Sartorius Stedim, Spain). Each filter was placed upside down on 1.5% non-nutrient agar (NNA) plates, containing Bacto Agar (DIFCO, USA), with Page’s amoeba saline seeded alongside heat-killed Escherichia coli (4). Following that, all the plates were sealed and incubated at 27°C to 29°C.
The gauze of dust samples was incubated on another NNA medium. Then, the plates were monitored in order to detect free-living amoeba plaques daily for 30 days. The positive samples were recultivated in order to harvest amoebae. The agar surface was scraped with a sterile swab after the addition of 5 mL of phosphate-buffered saline (PBS) to the medium. The suspension containing the amoebae was collected in a tube and washed 3 times with PBS (pH, 7.2) at 600 g for 2.5 minutes. As soon as the sediment contained adequate amoebae, it was transferred to a 1.5 microtube and kept at -20°C for DNA extraction.
3.5. DNA Extraction
Total genomic DNA from the positive samples was extracted using a Dyna Bio Kit (Takapuzist, Iran) and stored at -20°C.
3.6. Identification of Acanthamoeba Via PCR Assay
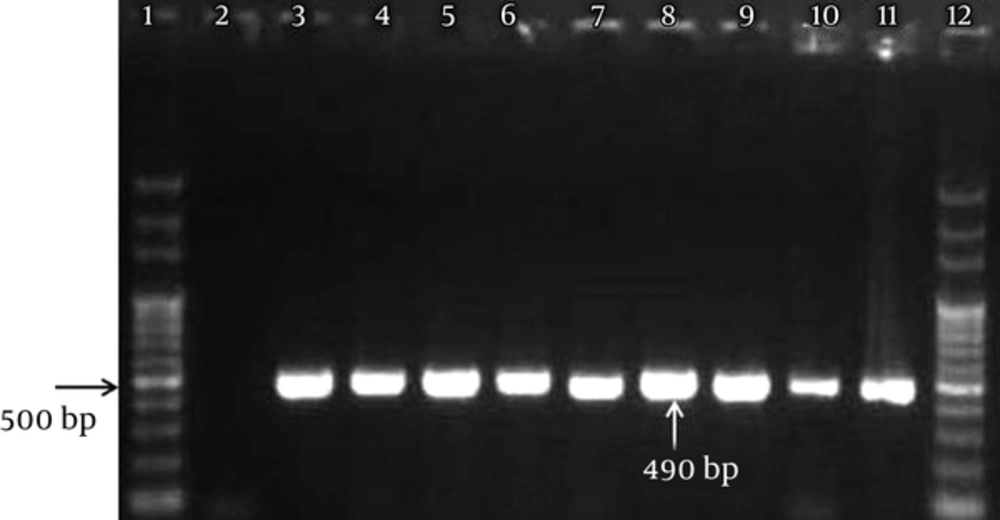
The PCR assay was performed using a pair of genus-specific primers, i.e., JDP1 (5′GGCCCAGATCGTTTACCGTGAA-3′) and JDP2 (5′TCTCACAAGCTGCTAGGGGAGTCA-3′), as described in the literature (4, 11). These sets of primers amplified a 500-bp fragment in 18S ribosomal RNA specific to Acanthamoeba species. The PCR reaction was performed in a volume of 20 μL, containing Taq DNA polymerase (Master Mix RED, Ampliqon, Denmark), 10 ng of DNA templates, and 10 pmol of each primer and distilled water. The PCR cycles were set up as follows: initial denaturation at 94°C for 4 minutes and 33 cycles of denaturation at 94°C for 35 seconds, annealing at 57°C for 45 seconds, extension at 72°C for 1 minute, and a final extension for 5 minutes at 72°C in a thermocycler (Flexcycler2, Germany). The PCR products were electrophoresed on 1.5% Agarose gel, stained with ethidium bromide solution (0.5 μg/mL), and visualized under UV light.
3.7. Sequencing
Twenty-nine positive samples (7 soil, 11 hospital dust, and 11 stagnant water samples) with perfect PCR products were selected for sequence analysis. Samples were sequenced in forward directions with a forward primer, JDP1, in an automatic sequencer (ABI 3730XL Genetic Analyzer, USA) at South Korea (via Takapozist, Iran). The genotype of Acanthamoeba spp. was identified by comparing each nucleotide sequence with DNA sequences from the GenBank. The Clustalw2 program was used for alignment of different sequences.
3.8. Statistical Analysis
The data were recorded in SPSS version 16.5 (SPSS Inc., Chicago, IL, USA) and analyzed by Chi square test. P value less than 0.05 was considered to be significant.
4. Results
In total, 122 environmental samples, including soil, hospital dust, and stagnant water samples, were collected in this study. Out of 122 environmental samples, 101 (82.8%) were positive for free-living amoebae in cultivation, as shown in Table 1. Based on the molecular method (PCR assay), 490-bp bands were identified as Acanthamoeba spp., using specific primers, JDP1 and JDP2 (Figure 1). Overall, 66 out of 122 (54.1%) environmental samples were positive for Acanthamoeba. The prevalence of Acanthamoeba spp. was 20 (62.5%) in soil, 21 (52.5%) in hospital dust, and 25 (50%) in stagnant water samples, respectively (Table 1); the difference was not statistically significant (P = 0.52). T4 was the most common Acanthamoeba genotype (76%), followed by T5 (13.8%). T2 (3.4%), T7 (3.4%), and T11 (3.4%) genotypes were also identified in soil and hospital dust samples (Table 2).
| Sources | Free-Living Amoeba | Acanthamoeba spp. | Total No (%) | ||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| Soil | 29 (90.6) | 3 (9.4) | 20 (62.5) | 12 (37.5) | 32 (100) |
| Dust | 29 (72.5) | 11 (27.5) | 21 (52.5) | 19 (47. 5) | 40 (100) |
| Stagnant water | 43 (86) | 7 (14) | 25 (50) | 25 (50) | 50 (100) |
| Total | 101 (82.8) | 21 (17.2) | 66 (54.1) | 56 (45.9) | 122 (100) |
| P Value | 0.095 | 0.525 | |||
aValues are expressed as No. (%).
| Sources/Genotypes | T4 | T5 | T2 | T7 | T11 | Total | Accession No. |
|---|---|---|---|---|---|---|---|
| Soil | 4 (57.1) | 2 (28.6) | 0 | 0 | 1 (14.3) | 7 (100) | LC179879, LC179880, LC276836-38, LC310895.1, LC310896.1 |
| Hospital dust | 7 (63.6) | 2 (18.2) | 1 (9.1) | 1 (9.1) | 0 | 11 (100) | LC184513-19, LC183867, LC276839-40, LC310897.1 |
| Stagnant water | 11 (100) | 0 | 0 | 0 | 0 | 11 (100) | LC177106-09, LC177665-67, LC179878, LC179877, LC276880-81 |
| Total | 22 (76) | 4 (13.8) | 1 (3.4) | 1 (3.4) | 1 (3.4) | 29 (100) |
aValues are expressed as No. (%).
The sequences showed 95% - 100% similarity to the deposited sequences in GenBank from other parts of Iran and other countries. Twenty-nine sequences were deposited in the GenBank, with the following accession numbers: hospital dust (LC184513-19, LC183867, LC276839-40, and LC310897.1), soil (LC179879, LC179880, LC276836-38, LC310895.1, and LC310896.1), and stagnant water (LC177106-09, LC177665-67, LC179878, LC179877, and LC276880-81).
5. Discussion
The current study showed that the prevalence rates of free-living amoebae and Acanthamoeba spp. were 90.6% and 62.5% in the soil samples, respectively. In a previous study, the free-living amoebae and Acanthamoeba spp. were identified in 26.9% and 17.3% of soil samples, respectively (8). In addition, the prevalence of Acanthamoeba has been estimated at 26% - 51.8% (6, 14, 15), which is lower than the results of the present study. On the other hand, Reyes-Batlle et al. (16) reported that 62.5% of soil samples from Gran Canaria, Spain were positive for Acanthamoeba spp., which is similar to our results. Overall, due to the high prevalence of Acanthamoeba spp. in soil, children are at a high risk of infection due to playing in parks.
In the present study, the prevalence of free-living amoebae and Acanthamoeba spp. in hospital dust from ENT, chemotherapy, and hemodialysis wards was 72.5% and 52.5%, respectively. The lowest rate of Acanthamoeba pollution was reported in the hemodialysis ward in the center of the city, while the highest rates were found in the ENT and chemotherapy wards, which lacked adequate health facilities. Also, water purification devices and sterilization/disinfection of hemodialysis ward were performed daily.
In previous studies, the prevalence of free-living amoebae in the immunodeficiency wards of hospitals was reported to be 52.9% (17) and 42.8% (18) in Tehran, Iran, respectively. On the other hand, the prevalence of Acanthamoeba spp. in hospital dust samples from Brazil was reported to be 23% (19), which is lower than the present study. Variations in geographic conditions, climate, and health facilities affect the incidence of Acanthamoeba. Kashan is a city situated near the desert. This region has low levels of rainfall and seasonal wind storms, which are followed by extreme dust storms. These conditions facilitate an increase in Acanthamoeba pollution and can explain the high prevalence of this parasite in our study.
The results of the present study showed that stagnant water samples were highly polluted with free-living amoebae. This finding is similar to the results of previous studies (80% - 88%) (10, 20). Moreover, in the current study, the prevalence of Acanthamoeba spp. in stagnant water samples was 50%, which is higher than the rates reported by other researchers from Iran (43%) (20) and Turkey (21%) (21). Furthermore, the results of sequencing revealed that most Acanthamoeba isolates belonged to T4 genotypes, followed by T5. In the present study, T4, T5, and T11 genotypes were identified in the soil. Another researcher has reported T3, T4, T5, and T11 genotypes in the soil of Iran (14). Reyes-Batlle et al. also found T4, T2, and T11 genotypes in soil samples from Spain (22).
In the present study, T4, T5, T2, and T7 genotypes were isolated from hospital dust samples in Kashan. In addition, Niyyati found T4, T5, and T11 genotypes in hospital dust (9). All stagnant water samples belonged to T4 genotypes, which is in agreement with the results of previous studies (23, 24). Maghsood et al. also reported Acanthamoeba T4 and T2 genotypes in clinical isolates (25). Acanthamoeba spp. seems to be responsible for an increase in fatal diseases, such as amoebic encephalitis (26). Furthermore, Acanthamoeba T4 genotype is pathogenic and predominantly associated with amoebic keratitis (4, 27).
6. Conclusions
The results of this study showed that the prevalence of free-living amoebae and Acanthamoeba spp. in environmental sources, including soil, hospital dust, and stagnant water was higher in Kashan, compared to other areas of Iran and other countries. The most important Acanthamoeba genotype was T4 in the hospital dust samples of the immunodeficiency ward. This organism is pathogenic and responsible for amebic keratitis and GAE; therefore, health education and sanitation services are recommended for the prevention of these diseases.