1. Background
Helicobacter pylori as microaerophilic and Gram-negative spiral shaped bacteria, are strongly associated with chronic gastritis (CG), peptic ulcer disease (PUD), and gastric cancer (GC) (1). In all patients, the infection can cause gastritis, yet only 10% to 15% will develop clinical signs (1, 2). Binding of H. pylori to the gastric epithelia, can result in the colonization, evasion from the immune system, and transfer of virulence factors to the gastric cells.
Helicobacter pylori outer membrane proteins (OMPs) can play an important role in binding to the gastric epithelia (1). About 4% of the genome is responsible for encoding OMPs, including SabA, AlpA and B, BabA, OipA, and HopQ (OMP 27) (1, 3-6). HopQ is divided to two families, HopQI and HopQII, which have amino acid sequence homology between 61% and 72% (7). Although the function of this protein is unknown, it has been suggested that it is involved in organism colonization, adhesion, and progress of peptic ulceration (7-9). The distribution of hopQI and hopQII genes varies in different populations and geographic regions (10-12). The hopQ shows a high degree of genetic diversity and H. pylori hopQI is more common in patients with peptic ulcer. Moreover, HopQI and HopQII are mostly present in cag A+ strains (13). On the other hand, SabA is a sialic acid-binding protein and can mediate binding of H. pylori to gastric mucosa cells. Sialyl-Lewis x/a (sLeX and sLea) antigens act as adhesin receptors for the SabA protein.
Sialylated glycoconjugates are found in small amounts in the stomach of healthy individuals yet in the presence of H. pylori infection, the expression of these antigens, especially sLea, are associated with gastric malignancies in both developed and developing countries. This issue represents the important role of SabA in gastric atrophy and gastric cancer. Therefore, more studies on SabA, particularly in developing countries, is suggested (14, 15). The association of different genotypes of H. pylori with clinical outcomes varies in different populations. Hence, investigation of putative virulence factors is crucial to provide useful clinical data in medical practice.
2. Objectives
Since there have been limited published epidemiologic studies on the prevalence of hopQ and sabA in H. pylori strains isolated from patients with gastrointestinal disorders in Iran, the present study aimed at evaluating the frequency of these genes among H. pylori isolates from patients with gastroduodenal disorders in an Iranian symptomatic population.
3. Methods
3.1. Ethics Statement
This study was approved by the Ethics Committee of Shahrekord University of Medical Sciences (EC = 92-11-31).
3.2. Patients, Endoscopy and Biopsies
In this cross-sectional study, conducted between March 2013 and April 2015, 150 H. pylori-positive patients, who underwent upper gastrointestinal endoscopy for a variety of indications at the Endoscopy Services of Hajar Hospital in Shahrekord, center of Iran, were included. Based on endoscopic and histological findings, the participants were classified to chronic gastritis (CG = 85), peptic ulcer disease (PUD = 61), and gastric cancer (GC = 4) groups. Some samples were taken from previously performed studies (16, 17). Written informed consent was obtained from all patients. Patients, who had undergone anti-H. pylori eradication therapy within the past three months, were excluded. During gastroscopy, biopsy samples were taken from the corpus for rapid urease test (RUT) and DNA extraction.
3.3. Rapid Urease Test (RUT)
For each patient, corpus samples were taken and tested for the presence of urease enzyme. The campylobacter-like organism (CLO) test (Romania) was used in this study. After 10 to 15 minutes at room temperature, if H. pylori urease was present in the tissue sample, it changed the gel from yellow to bright magenta.
3.4. Genomic DNA Extraction and Polymerase Chain Reaction (PCR)
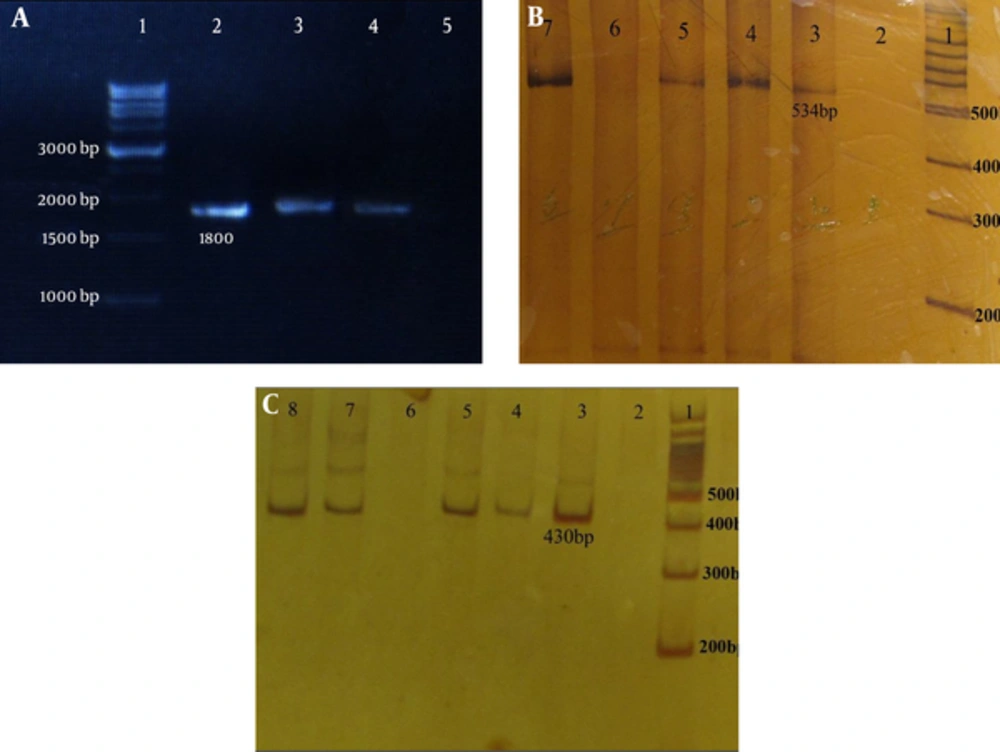
The phosphate buffer saline (pH = 7.2) microtubes containing corpus biopsies for DNA extraction were sent to the Cellular and Molecular Center, Shahrekord University of Medical Sciences. Genomic DNA was extracted using the DNA extraction kit (BioFlux Co., Japan), according to the manufacturer’s instructions. For H. pylori, molecular diagnosis at the species level (glmM and 16S rRNA), and for the presence of hopQI, hopQII and sabA genes, PCR assays were performed by previously described primers (7, 18, 19). Each PCR amplification was performed in a reaction volume of 25 µL, containing 1 × PCR buffer, 200 ng DNA, 4 mM MgCl2, 1.6 mM dNTP, 0.02 U Taq DNA polymerase and 0.16 pM primer (for hopQI) and 300 ng DNA, 4 mM MgCl2, 1.6 mM dNTP, 0.02 U Taq DNA polymerase, and 0.2 pM primer (for hopQII). To detect sabA, 200 ng DNA, 6 mM MgCl2, 3.2 mM dNTP, 0.02 unit Taq DNA polymerase, and 0.2 pM primer was used. The amplification of individual genes was done in a thermocycler (Corbett Research, Australia), as described previously (10, 19, 20). Amplicons were analyzed on 1% agarose gel and stained by the ethidium bromide (0.5 µg/mL) for sabA and 6% polyacrylamide gel with TBE 5× for hopQI and hopQII genes, and examined under UV illumination (Figure 1).
Agarose gel electrophoresis of sabA and polyacrylamid gel of hopQI and hopQII genes in H. pylori isolates. A, H. pylori sabA: lane 1: 1kb DNA ladder marker, lanes 2 - 4 (1800 bp): sabA positive strains, and lane 5 is negative for sabA. B, H. pylori hopQI: lane 1: 100 bp DNA ladder marker, lanes 3 - 5 and 7 (525 bp): hopQI positive strains and lanes 2 and 6 are negative for hop QI. C, H. pylori hopQII: lane 1: 100 bp DNA ladder marker, lanes 3 - 5 and 7 - 8 (430 bp): hopQII positive strains, and lanes 2 and 6 are negative for hopQII.
3.5. Data Analysis
The Chi-square (χ2) and Fisher’s exact tests were used for analysis of significant differences between the studied virulence genes with the clinical outcome results using the SPSS software (version 22.0; IBM Co., Armonk, NY, USA). The results of demographic and clinical manifestations were presented as descriptive statistics in terms of relative frequency. P values of < 0.05 were considered statistically significant.
4. Results
In the current study, 150 H. pylori strains obtained from patients (85 females and 65 males, ranging in age from 15 to 85 years with a mean age of 43 years, which were categorized to three groups, including patients < 30, between 30 and 50, and > 50 years old) with gastroduodenal disorders after confirmation by RUT and PCR (for detection of glmM and 16S rRNA genes), were evaluated for the presence of hopQI, hopQII and sabA genes. The hopQI, hopQII, and sabA genes were detected in 74 (49.3%), 59 (39.3%), and 43 (28.7%) of the isolates, respectively (P > 0.05). The distribution of these genes in CG, PUD, and GC cases, were as follows: hopQI (42.4%, 57.4%, and 75%), hopQII (38.8%, 39.3%, and 50%) and sabA (29.4%, 26.2%, and 50%). No significant difference (P > 0.05) was observed between age or gender and clinical outcome (Tables 1 and 2). The association between disease type and the presence of hopQI, hopQII and sabA genes was also not significant (P > 0.05).
| Gender | Disease Group | Total | |
|---|---|---|---|
| Group 1 | Group 2 | ||
| Female | 29 (44.6) | 56 (65.9) | 85 (56.7) |
| Male | 36 (55.4) | 29 (34.1) | 65 (43.3) |
| Total | 65 (100) | 85 (100) | 150 (100) |
aValues are expressed as No. (%).
bGroup 1: Patients with peptic ulcer disease and gastric cancer, group 2: patients with chronic gastritis.
| Age | Disease Group | Total | |
|---|---|---|---|
| Group 1 | Group 2 | ||
| Group 1 | 17 (26.2) | 18 (21.2) | 35 (23.3) |
| Group 2 | 31 (47.7) | 45 (52.9) | 76 (50.7) |
| Group 3 | 17 (26.2) | 22 (25.9) | 39 (26) |
aValues are expressed as No. (%).
bAge group 1: patients less than 30 years old, age group 2: patients between 30 to 50 years old, age group 3: patients more than 50 years old, disease group 1: patients with peptic ulcer disease and gastric cancer, disease group 2: patients with chronic gastritis.
5. Discussion
The present study evaluated the frequencies of H. pylori hopQI, hopQII, and sabA genes among patients with gastroduodenal diseases in Shahrekord, Iran. The hopQI gene was predominant (49.3%) in comparison with hopQII and sabA genes with frequencies of 39.3% and 28.7%, respectively. Host’s genetics, environmental factors, such as diet, and H. pylori virulence determinants are associated with the severity of clinical outcomes (21, 22). Genotypic analysis of H. pylori is also a useful method in screening of gastric diseases (20). In the recent years, the role of H. pylori OMPs, such as hopQ and sabA, in developing of digestive diseases have been investigated. Therefore, the association of virulence genes with clinical outcome needs to be further investigated in clinical practice. It has shown that hopQ can be a candidate adhesin to gastric epithelium cells (8).
The frequencies of hopQI and II alleles vary depending on geographic regions. Previous studies in Western and Eastern countries have shown that there is a higher prevalence of hopQI than hopQII. The prevalence of hopQI and hopQII genes in the United States and East Asian countries were reported as 55.5%, 30.9% and 88.8%, 0.7%, respectively (10). Moreover, this frequency for hopQI among Colombian and Italian patients was reported as 72% and 29%, respectively (11, 12). The current results (49.3% for hopQI and 39.3% for hopQII) are similar with a study conducted in the United States (10). Indeed, in the study of Ohno et al. (9) among Asian and Western population, no significant association was observed between hopQ genotypes and clinical outcome. In the first ever study conducted in Iran, the frequency of hopQI (33%) was mentioned lower than hopQII (52%), and the hopQII genotype was found to be associated with gastric cancer (20). The reported results are in contrast with the current study, so that no association was found between gastrointestinal diseases and hopQ alleles (P > 0.05). The current findings are consistent with the results of two other studies from Iran (23, 24).
Due to the absence of sLeX in normal gastric epithelia, the sabA can be related to the H. pylori chronic persistence and severe clinical outcomes, such as gastric cancer (14, 25). The present data (28.7%) compared to studies in Taiwan (80%), France (86%), and Netherland (93%), revealed a much lower rate (26-28). The sabA frequency among Japanese patients from different population and times, was also found to be 81% and 91%, respectively (29, 30). Yamaoka et al. in the United States reported that sabA frequency in patients with gastritis, duodenal ulcer and gastric cancer was 66%, 44%, and 70%, respectively, and a significant association was established between sabA and gastritis and gastric cancer (31).
In three previous studies from Iran, the frequency of sabA was reported as 86.6%, 83.6%, and 100%, respectively, with no association with the severity of clinical outcomes (32-34), and this was in line with the current findings and another study from Japan (29). In comparison with the above-mentioned studies from different countries, the current results showed a considerably lower rate of sabA. This issue highlights that sabA genetically varies among different strains and geographic areas (30), and might not be present in all H. pylori isolates (14). On the other hand, to establish an association between H. pylori virulence determinants and clinical outcome, patient selection is crucial because the studied participants should be sufficiently large and heterogeneous (35).
5.1. Conclusions
Consistent with most other studies, the rate of hopQI in the current study was higher than hopQII; however, sabA frequency in contrast to other reports was considerably low. Taken together, regarding the heterogeneity of H. pylori isolates and diverse results from different geographic areas, further intensive studies are needed to assess the role of the above-mentioned genes in H. pylori pathogenesis and their association with clinical outcomes in Iran.