1. Background
The search for isolating novel antibiotics, effective against resistant pathogenic microorganisms from unexplored habitats around the world, continues to be an important sector of research. There is an emerging crisis secondary to the development of antibiotic resistant microbial pathogens and toxicity of several antibiotics in use (1, 2). As a result of the extensive screening, about 17000 antibiotics from different microbial sources have been isolated (3). The role of actinomycetes, as a source of bioactive molecule compounds, are well known, as these organisms, especially Streptomyces, provides more than half (70%) of the naturally occurring antibiotics extensively used by the pharmaceutical industry (3, 4).
During the past two decades, there has been a decline in the discovery of new compounds from soil-derived actinomycetes (5). Hence, in the quest for exploring newer ecosystems for the isolation of actinomycetes and to discover high efficacy antibiotics, it is crucial to overcome the problem of antibiotic resistance. The Western Ghats region of India is being explored for its rich and varied heritage of microbial diversity. Several strains of actinomycetes with antimicrobial activity have been isolated and characterized from soil samples of Coimbatore, Kerala, Tamil Nadu, India (6-10). However, there is a lack of adequate data on isolated actinomycetes strains with potential antibiotic properties from the Kodagu region of Karnataka.
2. Objectives
The objective of the present study was to isolate, identify and evaluate the antimicrobial activity for optimization of the strain Streptomyces obtained from a soil sample of Kodagu district of Karnataka, India.
3. Materials and Methods
3.1. Sample Collection
Soil samples were collected randomly from agricultural lands of Kushalnagar Taluk, Kodagu district, Karnataka, India during 2011 using an open-end soil borer (20 cm in depth and 25 cm in diameter) from a depth of 10 - 20 cm, which were then air dried for 24 to 48 hours (11). The soil sample was then treated with 1% CaCO3 and kept at ambient temperature for 2 - 3 days (12) for the next step of analysis.
3.2. Isolation of Actinomycetes
Soil samples were serially diluted (13) and plated on Actinomycetes Isolation Agar medium procured from Difco (Hi media, Mumbai, India) and incubated at 28˚C for 6 days. The suspected colonies were picked up and purified on yeast extract-malt extract agar medium (ISP-2) supplemented with 50 µg/mL Nystatin, as antifungal agent (14). The pure culture of ACTK2 was maintained on ISP-2 slants at 4˚C. The spore mass and mycelium of the pure isolate were stored at -20˚C as glycerol suspension (20%, v/v) for further analysis (Difco, Hi media, Mumbai, India) (15).
3.3. Taxonomical Characterization of the Strain ACTK2
The strain of ACTK2 was identified based on cultural, morphological, physiological, biochemical and sequence analysis of 16S rRNA gene parameters.
3.3.1. Cultural and Morphological Characteristics
The cultural characteristic of pure isolate was studied after incubation of strain ACTK2 for 7-14 days at 28˚C using various media: Tryptone Yeast Extract Agar (ISP-1), Yeast Extract-Malt Extract Agar (ISP-2), Oat Meal Agar (ISP-3), Starch-Casein nitrate Agar medium (ISP-4), Glycerol-Asparagine Agar (ISP-5), Peptone Yeast Extract Iron Agar (ISP-6), Modified ISP-2 (MISP-2), Starch Agar, Potato Dextrose Agar, Czapek Dox Agar, Nutrient Agar, Modified Nutrient Glucose Agar (MNGA) and Mueller Hinton Agar, procured from Hi-Media, Mumbai, India. Morphological parameters such as aerial and substrate mycelium, arrangements of spores and Gram staining reaction of the ACTK2 strain (16) were carried out as described in Bergy's Manual of Systematic Bacteriology and by Shirling and Gottlieb (17, 18).
3.3.2. Physiological and Biochemical Characteristics
Various tests, such as the catalase test, citrate utilization test, nitrate reduction test, starch hydrolysis (19), gelatin hydrolysis (20) and production of melanin pigmentation on ISP-6 (18), using standard media from Hi media, Mumbai, India, were carried out to analyze the physiological and biochemical characteristics of ACTK2. The analysis of 2-6 diaminopimelic acid (DAP) isomers, present in the whole cell wall composition of the strain ACTK2, was determined by thin layer chromatography (S D Fine-Chem Ltd., Mumbai, India) (21).
3.3.3. Molecular Identification of ACTK2
The isolated ACTK2 was grown by agitation in 500 mL flasks containing 100 mL of ISP-2 medium for 4 days at 28˚C. After the incubation, biomass was harvested by centrifugation at 10000 rpm for 5 minutes and washed twice with double-distilled water. The genomic DNA of ACTK2 was isolated using the AMpurE Bacterial Genomic DNA Mini kit (Amnion’s Bioscience, Bangalore, India) (22). The formed DNA pellet was air dried and dissolved in 100 μL of TE buffer (100 mM NaCl, 1 mM EDTA, 100 mM Tris-HCl, pH 8.0) and used for polymerase chain reaction (PCR) studies. The 16S ribosomal RNA gene was amplified by using PCR with Taq DNA polymerase and universal primer pairs 27F (5´-AGAGTTTGATCCTGGCTCAG-3´) and 1492R (5´-GGTTACCTTGTTACGACTT-3´) (23).
Amplification was performed using Thermal Cycler (Peqlab Biotechnologie GmbH, Erlangen, Germany), in a total volume of 50 μl containing 50 ng/mL DNA, 100 pmol of each primer, 10 mM dNTPs, 2 μL10X Taq Polymerase buffer and 0.25 U Taq DNA polymerase (Amnion's PCR Master-Mix kit, Amnion’s Bioscience, Bangalore, India). The PCR conditions included, denaturation at 94˚C for 5 minutes followed by 35 cycles of denaturation at 94˚C for 30 seconds, primer annealing at 55˚C for 30 seconds and primer extension at 72˚C for 1.5 minutes and final extension at 72˚C for 5 minutes. The PCR products were detected by 1.0% (w/v) agarose gel containing ethidium bromide (0.5 µg/mL) and were visualized by the ultraviolet (UV) fluorescence gel documentation system (UTP-Bio Doc, USA).
3.3.3.1. Cloning and Sequencing
The PCR products were cloned in the pGEM-T Easy Vector (Promega Corp., Fitchburg, Wisconsin, USA) and transformed to Escherichia coli cells for nucleotide sequence determination using an automated sequencer (Genetic Analyzer 3130, Applied Biosystems, Waltham, Massachusetts, USA). The nucleotide sequences analysis of the 16S rRNA gene of ACTK2 was compared with the reference species of bacteria contained in the GenBank database, using the NCBI BLAST available at http://blast.ncbi.nlm.nih.gov website.
3.3.3.2. Phylogenetic Analysis of Isolate ACTK2
The phylogenetic tree was derived from the distance matrixes using the neighbor-joining method (24). All the analyses were performed on a bootstrap dataset containing 100 replicates (generated by the program) (24).
3.4. Antimicrobial Activity
The test organisms employed included the following: Gram-positive Staphylococcus aureus (MTCC 96), Bacillus subtilis (MTCC 121) and Gram-negative E. coli (MTCC 729), Enterococcus aerogenes (MTCC 2829). All the test organisms used in this study were obtained from the Microbial Type Culture Collection (MTCC), Institute of Microbial Technology (IMTECH), Chandigarh (India). The filamentous fungi Trichoderma harizianum (MTCC6046) and Fusarium proliferatum (MTCC 9375) were obtained from the Department of Studies in Microbiology and Department of Studies in Botany, University of Mysore, Mysore, Karnataka, India. Primary screening of antimicrobial activity of Streptomycesflavogriseus (ACTK2) was determined by using the perpendicular streak technique (25). Colonies were streaked on the center of ISP-2 plates and incubated at 28˚C for 4 - 5 days. The test organism was inoculated perpendicular to the strain ACTK2 and the plate was incubated at 37˚C for 24 hours for bacteria and at 28˚C for 96 hours for fungi (8). The zone of inhibition was determined using a millimeter scale.
3.5. Optimization of Fermentation Parameters
To determine the optimal nutritional and cultural conditions for the growth and formation of antimicrobial compound by the ACTK2 strain using different media, such as ISP-2 Broth, Starch Casein Broth, MISP-2 Broth, ISP-4 Broth and MNG Broth were used. Culture conditions at different pH values (4, 6, 8 and 10), temperatures (20, 28, 37 and 45ºC) and incubation periods (3, 4, 5, 7, 10 and 15 days) were studied for microbial growth and production of antimicrobial compound. Fermentation was carried out in an 250 mL Erlenmeyer flask, containing 50 mL of medium inoculated with 100 µL of the spore suspension of the strain ACTK2 (1.5 × 105 CFU/ µL) and incubated in an orbital shaker at 100 rpm (26).
3.6. Determination of dry Weight of Mycelium
Growth of the isolate ACTK2 was measured as dry weight of the mycelium after 3, 4, 5, 7, 10 and 15 days of incubation. The biomass was separated after centrifugation at 4000 rpm for 10 minutes and transferred to a pre-weighted dry filter paper using a clean spatula and was kept in an oven at 60˚C for 12 hours to reach a constant weight. The amount of growth was expressed as mg/50 mL culture medium (27).
3.7. Extraction of Antimicrobial Compound From ACTK2
The antimicrobial compound was extracted from the mycelium-free supernatant by the solvent-liquid extraction method (28). N-butanol was added to the supernatant in the ratio of 1:1(v/v) and shaken vigorously. The butanol phase having antimicrobial compound was separated and the extraction process was repeated three times. The n-butanol phase was concentrated by using a rotary vacuum flash evaporator at 60˚C, 200 rpm. The obtained residue (crude extract) was dissolved in 1 mL of n-butanol solvent and stored at 4˚C.
3.7.1. Determination of the Antimicrobial Activity
The antimicrobial activity of the n-butanol extract of ACTK2 was determined by the disc diffusion method (29) against the test organisms. The amount of 20 mL of sterilized molten Muller Hinton Agar was seeded with 50 µL of test organisms (1.5 × 105 CFU/µL), swirled gently and aseptically poured into Petri dishes and allowed to solidify. Sterile filter paper disc containing 50 µL of the crude solvent extract was placed at the center of the Petri dish. Control was maintained by loading 50 mL of n-butanol in place of crude extract. The plates were kept at 4˚C for 2 hours for diffusion of the antibiotic and then incubated at 37˚C for bacteria and at 28˚C for 48 hours for fungi. The inhibition zone around the disk was measured with a millimeter scale and the results were tabulated and analyzed.
4. Results
The aerobic actinomycete designated as strain ACTK2 (accession number KC990785) isolated from the soil sample of Kushalnagar Taluk, Kodagu district, Karnataka, India, was a Gram-positive, S.flavogriseus. The culture was a slow growing while the filamentous aerial mycelium was grayish to whitish yellow in color producing straight to flexuous chains of spores. The sporulation on the agar medium was observed after 2 - 3 days of incubation (Table 1). No diffusible pigment was detected on the used media, except for ISP-3 medium. These morphological characteristics strongly suggested that the ACTK2 strain belonged to the genus Streptomyces. The physiological and biochemical characteristics of ACTK2 are presented in Table 2. The ACTK2 hydrolyzed Starch, reduced Nitrate, liquefied Gelatin, but it did not hydrolyze Citrate and produced Hydrogen Sulphide (H2S). The ACTK2 strain was Catalase positive and Oxidase negative. Melanin production was not observed on ISP-6. The cell wall composition of the strain ACTK2 was found to be L-DAP (cell wall chemotype-1) which had characteristics of the genus Streptomyces.
| Sl. No. | Medium | Growth | Aerial Mycelium | Substrate Mycelium | Spores | Soluble Pigment |
|---|---|---|---|---|---|---|
| 1 | ISP-1 | Good | Moderate, Ashes | Dark brown | Ashes | None |
| 2 | ISP-2 | Good | Abundant, Ashes | Dark brown | Ashes | None |
| 3 | ISP-3 | Good | Abundant, Gray | Black | Gray | brown |
| 4 | ISP-4 | Good | Abundant, Gray | Black | Gray | Light brown |
| 5 | ISP-5 | Good | Moderate, Whitish gray | Whitish yellow | Grayish white | Light brown |
| 6 | ISP-6 | Poor | None | Yellowish brown | None | Medium yellow brown |
| 7 | MNGA | Moderate | Poor, Light gray | Whitish gray | Whitish gray | None |
| 8 | NA | Moderate | Poor, Light gray | Whitish gray | Gray | None |
| 9 | MHA | Moderate | Moderate, Grayish white | Whitish gray | Gray | None |
| 10 | PDA | Moderate | Poor, Creamish yellow | Whitish yellow | Whitish gray | None |
| 11 | Starch Agar | Moderate | poor ,Whitish yellow | Whitish yellow | Whitish gray | None |
| 12 | CZP | Good | Abundant, gray | Dark brown | gray | Brownish black |
| 13 | MISP-2 | Good | Abundant, ashes | Dark brown | ashes | Light brown |
aAbbreviations: ISP-1-6: International Streptomyces Project medium; MNGA: Yeast Extract- Malt Extract Dextrose Agar; NA: Nutrient Agar; MHA: Muller-Hinton Agar; PDA: Potato Dextrose Agar; CZP: Czapek Dox Agar; MISP-2: Modified Yeast Extract-Malt Extract Agar.
| Sl. No, Properties | Streptomyces flavogriseus ACTK2 |
|---|---|
| I. Morphological characteristics | |
| 1. Sporophore morphology | Flexible spiral |
| 2. Color of aerial mycelium | Ashes gray |
| 3. Color of substrate mycelium | Dark gray |
| 4. Spore mass | Ashes gray |
| II. Biochemical characteristics | |
| 1. Catalase test | + |
| 2. Oxidase test | - |
| 3. Citrate utilization | - |
| 4. Gelatin hydrolysis | + |
| 5. Nitrate reduction | + |
| 6. Starch hydrolysis | + |
| 7. Urease test | + |
| 8. Melanin production | - |
| III. Chemotaxonomic characters | |
| 1. Cell wall amino acid analysis | L-DAP |
| IV. Grams staining | + |
aAbbreviation: L-DAP, L- diaminopimelic acid.
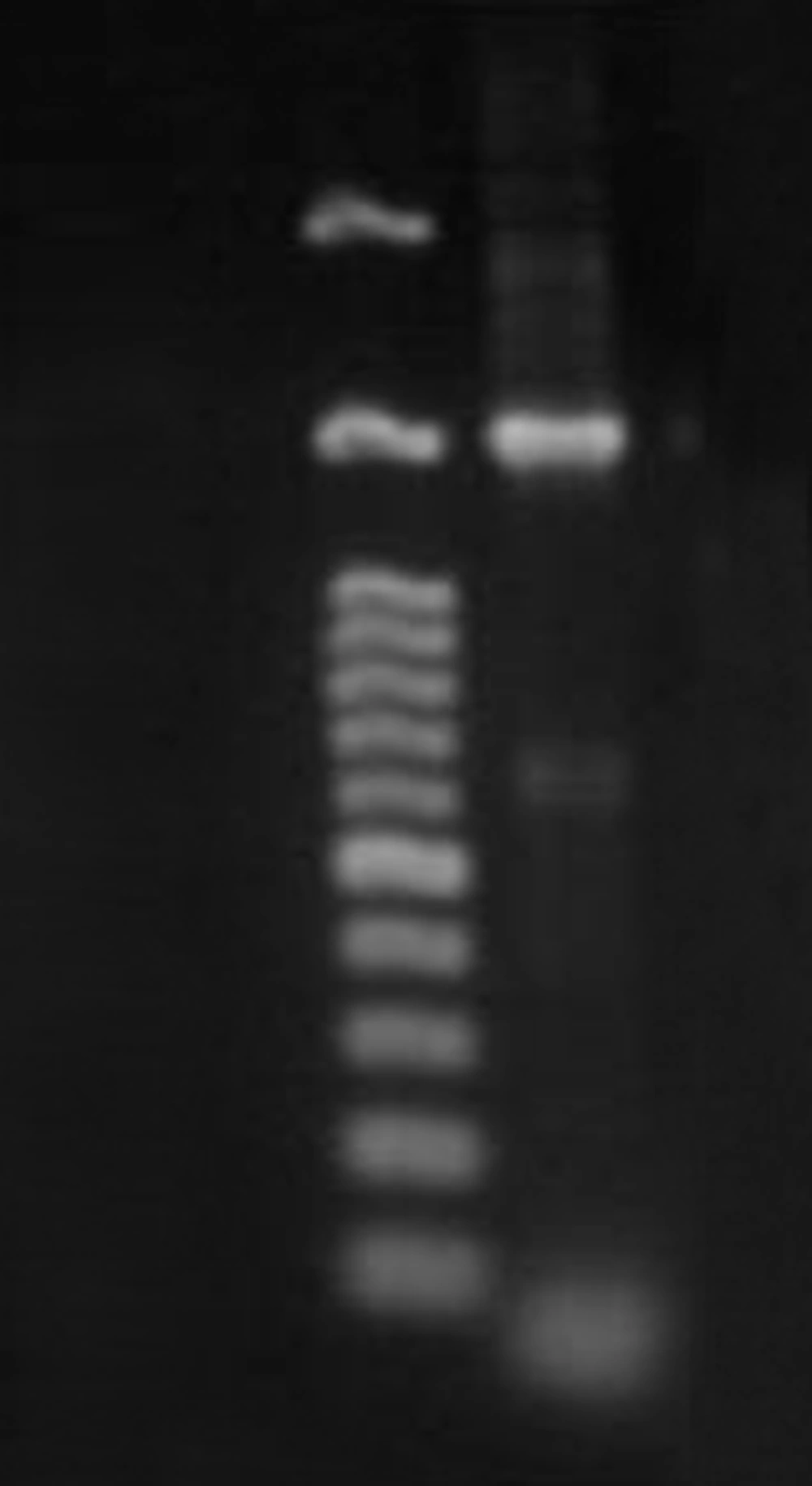
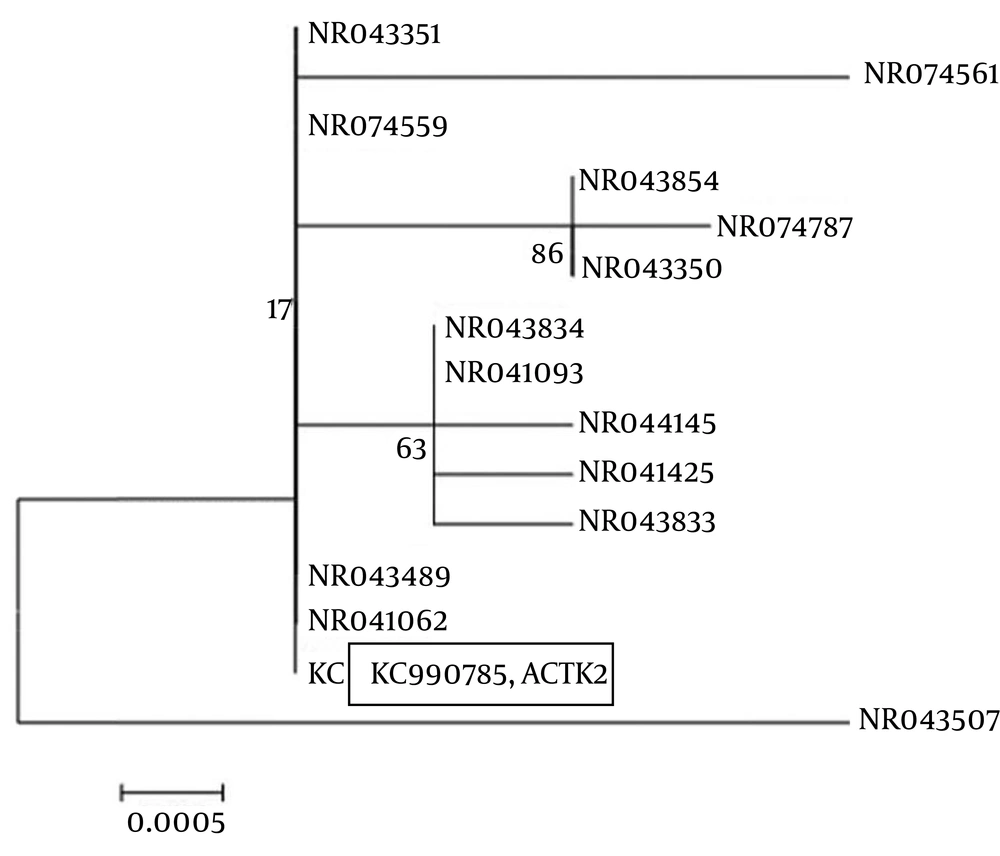
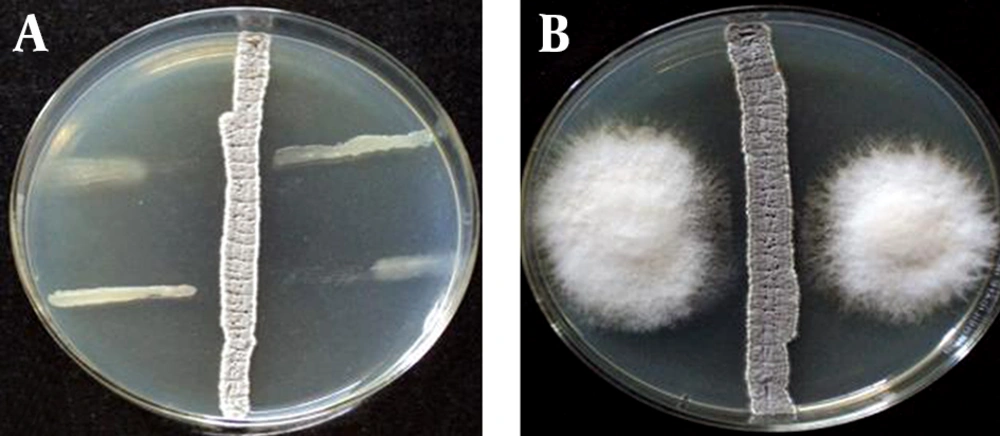
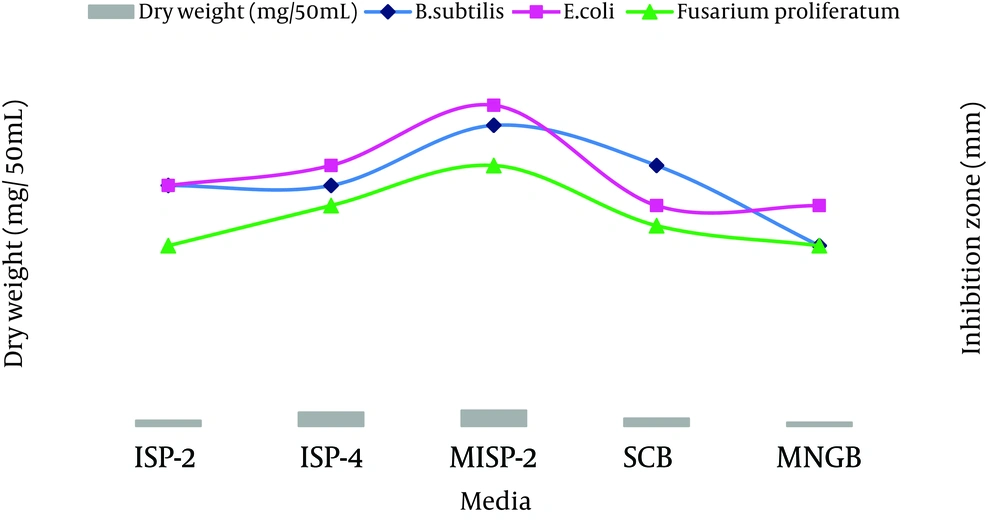
The results of the PCR and sequencing of the strain ACTK2 are presented in Figureس 1 and 2. The alignment of the nucleotide sequences (1489 bp) of strain ACTK2 in the gene bank using NCBI exhibited a similarity level of 100% with S. flavogriseus. The phylogenetic tree obtained by applying the neighbor joining method is shown in Figure 3. Primary screening of ACTK2 for its antimicrobial activity showed a broad spectrum antimicrobial activity against test bacteria and F. proliferatum. The strain ACTK2 did not show any activity against T. harizianum (Figure 4). The n-Butanol extraction of ACTK2 showed maximum inhibitory activity against E. coli followed by B. subtilis, S. aureus, F. proliferatum and S. aureus, respectively. Among the Gram-positive and Gram-negative bacteria, E. coli and B. subtilis were the most sensitive to antimicrobial compounds produced by ACTK2. The optimization studies revealed that ACTK2 was able to grow in all the tested culture media. However, the maximum growth (biomass) and production of antimicrobial compound was observed in MISP-2 Broth with an average dry weight of mycelium (0.8 mg/50 mL) followed by Inorganic salts-Starch Broth (ISP-4) (0.7 mg/50 mL), Starch Casein Broth (0.4 mg/50 mL), Yeast Extract- Malt Extract Glucose Broth (ISP-2) (0.3 mg/50 mL) and Modified Nutrient Glucose Broth (MNGB) (0.2 mg/50 mL), respectively (Figure 5).
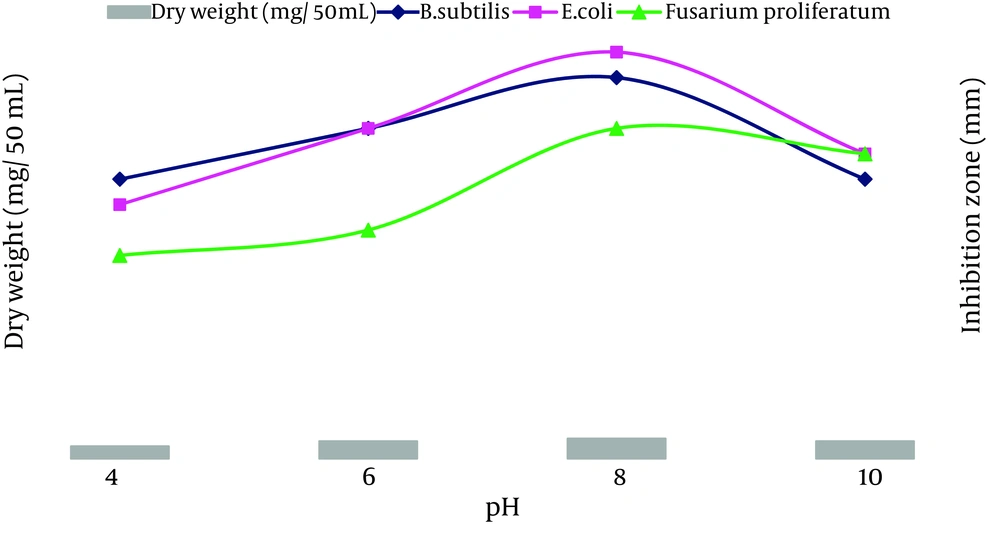
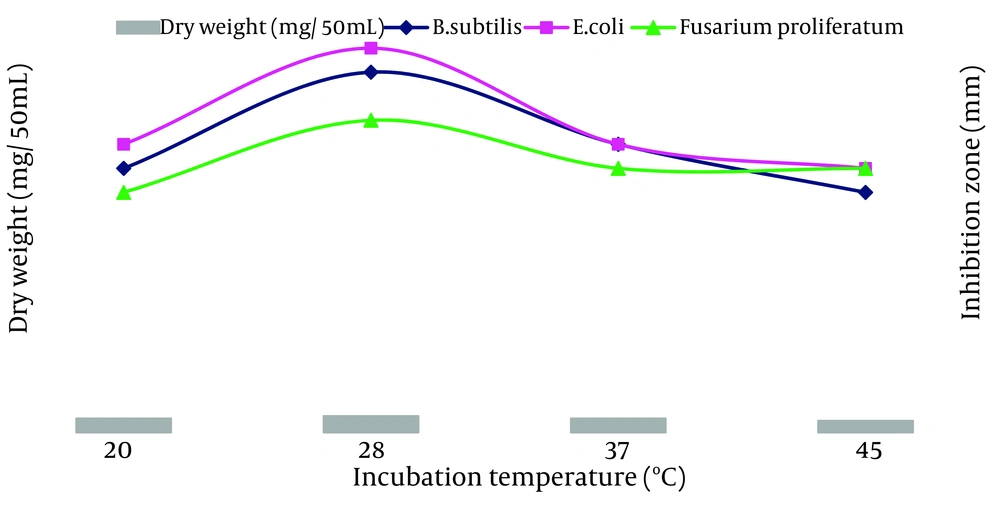
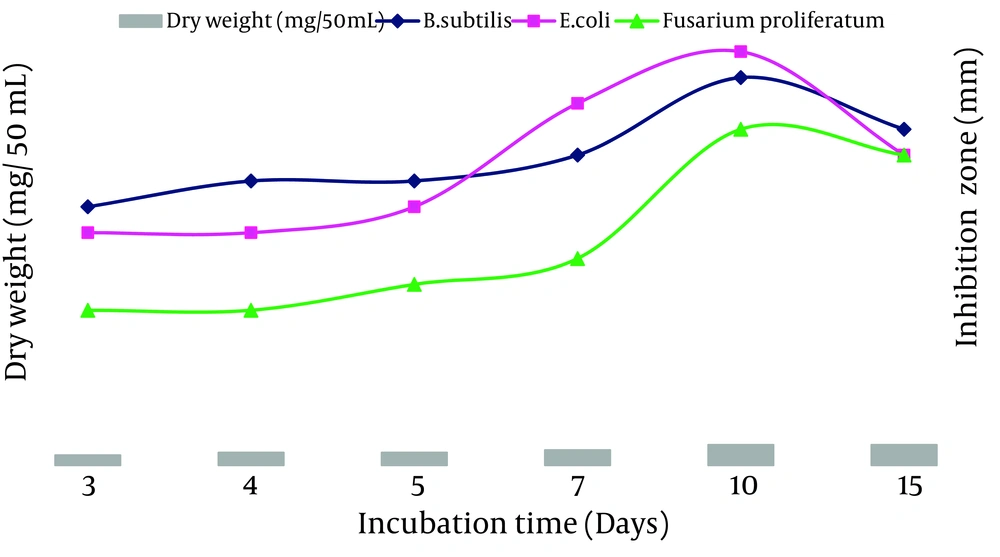
The ACTK2 could grow and produce the antimicrobial compound up to a pH 10.0. The maximum production was observed at a pH 8.0 (Figure 6). The results suggest that strain ACTK2 was slightly alkaliphilic in nature. The ACTK2 showed a narrow range of incubation temperature for relatively good growth and antimicrobial production. Higher growth (0.8 mg/50 mL), as well as antimicrobial activity, were noticed against B. subtilis at 28˚C (Figure 7), indicating that ACTK2 was mesosphilic in nature. An incubation period of up to 10 days was found to be optimal for maximum growth (0.8 mg/50 mL), as well as antimicrobial production (Figure 8). The results of optimization of nutritional and cultural conditions showed that n-butanol extract of strain ACTK2 exerted maximum activity against E. coli (16 mm), followed by B. subtilis (15 mm), F. proliferatum (13 mm), Staph. aureus (13 mm) and Staph. aureus (11 mm).
5. Discussion
The emerging crisis of antibiotic resistant pathogens indicates an increasing necessity for the survey of unexplored and underexplored niche habitats. The isolation of actinomycetes can contribute to the discovery of novel, safe, effective, broad-spectrum antimicrobial compounds, as part of the strategy to control the drug-resistant pathogens (30, 31). During the course of screening of bioactive compounds for the isolation of new antibiotics each year, thousands of actinomycetes, particularly Streptomyces strains, are screened by pharmaceutical research laboratories as sources for novel antimicrobial compounds (8, 27, 32-34). In the present study, a broad spectrum of antimicrobial producing actinomycetes, designated as S. flavogriseus (ACTK2) isolated from agricultural soil sample of Kushalnagar, Kodagu, Karnataka state, India were selected for optimization of their antimicrobial activities. The isolate ACTK2 was identified through conventional and molecular methods. The isolate exhibited broad antimicrobial activities against Gram-positive, Gram-negative bacteria and fungus F. proliferatum in the primary and secondary screening process. In a study conducted in Iran (35) with different strains of Streptomyces, there was no significant correlation between the activity of intact bacteria in primary screening and their extract in secondary screening, which was similar to the present study on ACTK2.
The results of the optimal nutritional media showed that antibiotic production was higher in the medium containing glucose. The importance of glucose in the nutritional medium for the synthesis of a wide range of antibiotics by different Streptomyces species has been reported by many investigators (27, 36-38). Although the strain ACTK2 was able to grow in five different media tested, the highest growth (0.8 mg/50 mL) and antimicrobial activity were obtained in MISP-2 Broth medium supplemented with starch (as a carbon source) and CaCO3 (as growth promoters). This result is quite comparable with the S. fulvissimus and Pseudonocardi species, for which MISP-2 was found to be a suitable medium for the antibiotic production (39, 40). Optimal cultural conditions at different pH levels and incubation temperatures and time for antimicrobial production by S. flavogriseus improved in the medium containing MISP-2 Broth.
The results revealed that the maximum growth (0.8 mg/50 mL) and in-vitro antimicrobial compound production by S. flavogriseus could be obtained in MISP-2 medium having a pH 8, for 10 days of incubation at 28˚C. The growth profile, closely coupled with the metabolic capacities of the produced organism S. flavogriseus, which greatly influenced the biosynthesis of antibiotics. The findings confirm that the nature of the medium composition and cultural conditions strongly affect and enhance the antimicrobial compound of ACTK2, which is comparable to the surveys conducted on different Streptomyces spp. by other investigators (9, 27, 32-34, 36). In this study, the S. flavogriseus designated as ACTK2, isolated from agricultural soil sample of Kushalnagar Taluk of Kodagu, Karnataka, India, showed broad spectrum antimicrobial activity against Gram-positive, Gram-negative bacteria and the fungus F. proliferatum. Further studies on the purification and chemical characterization of the antimicrobial compound from the strain of S. flavogriseus will be useful for pharmaceutical applications.