1. Background
Enteropathogenic Escherichia coli (EPEC) is a leading cause of infantile diarrhea in developing countries. These pathogens are characterized by their ability to cause attaching and effacing lesions in intestinal mucosa. The lesions are directed by a pathogenicity island (PAI) known as Locus of Enterocyte Effacement (LEE), which encodes a type III secretion apparatus, intimin and effector proteins. Typical EPEC (tEPEC) strains have fimbrial adhesion called bundle-forming pili (BFP) and maybe more pathogenic than atypical strains which do not possess it (1). In developing countries, in most cases, EPEC isolates recovered from humans with diarrhea are tEPEC; however, in industrial countries, most strains do not possess BFP and are identified as atypical EPEC (aEPEC) (2).
The spread of extended-spectrum β-lactamases (ESBLs) is an emerging global public health problem. Most ESBL genes are mutant derivatives of the classical blaSHV and blaTEM β-lactamases, but a rapid increase in the prevalence of blaCTX-M has been reported among Enterobacteriaceae over the last decade. These genes are capable of conferring resistance to third-generation cephalosporins (e.g. ceftazidime and cefotaxime) and aztreonam, but not cephamycins (e.g. cefoxitin) and carbapenems (3). In contrast to ESBLs, AmpC β-lactamases are poorly inhibited by clavulanic acid and are active against cephamycins (4). Antibiotic resistance is on the rise among diarrheagenic E. coli in developing countries, where overuse and misuse of antibiotics is common (5, 6). Furthermore, emergence of ESBL genes within commensal E. coli isolates in children is a matter of concern and has long been known that these bacteria are potential reservoirs for those genes in both community and hospital settings (7, 8). Therefore, information regarding ESBLs as well as other classes of β-lactamases in diarrheagenic pathogens should be considered in clinical management when an optimal treatment is needed (6). EPEC still plays an important role as a causative agent of infantile diarrhea in our country (9, 10). Although there were epidemiological surveys regarding prevalence of EPEC in Iran, none of these studies investigated the occurrence of β-lactamase genes in those isolates.
2. Objectives
Considering the paucity of epidemiological data on the issue, this study was designed to estimate the prevalence of blaCTX-M, blaSHV and blaTEM genes among EPEC strains obtained from children with diarrhea. The second objective was to determine the genetic diversity among ESBL producing strains.
3. Materials and Methods
3.1. Sampling and Detection of EPEC Strains
Stool specimens were obtained from children with diarrhea (≤ 10 years old) during 17 months (September 2011 to January 2013). The sample size was calculated using the following formula: N = Z2 pq/d2. Thus, a sample size (n) of 350 patients was sufficient, assuming that the prevalence (p) of EPEC in children was 9% (9, 10). The expected margin of errors (d) was 0.04 and the confidence interval (CI) was 95%. The patients were admitted to one of three pediatric hospitals in Tehran. They had evidence of more than three episodes of watery, loose or bloody stools per day. In Brief, all stool suspensions (one stool per patient) were inoculated directly onto MacConkey agar (Merck, Germany) plates and incubated at 37°C for 24 hours. For detection of E. coli strains, up to five lactose-positive colonies per plate were selected and subjected to routine biochemical tests (Gram staining, oxidase test, indole production, H2S production, carbohydrate utilization on TSI agar, MRVP reaction, urease production, etc.).
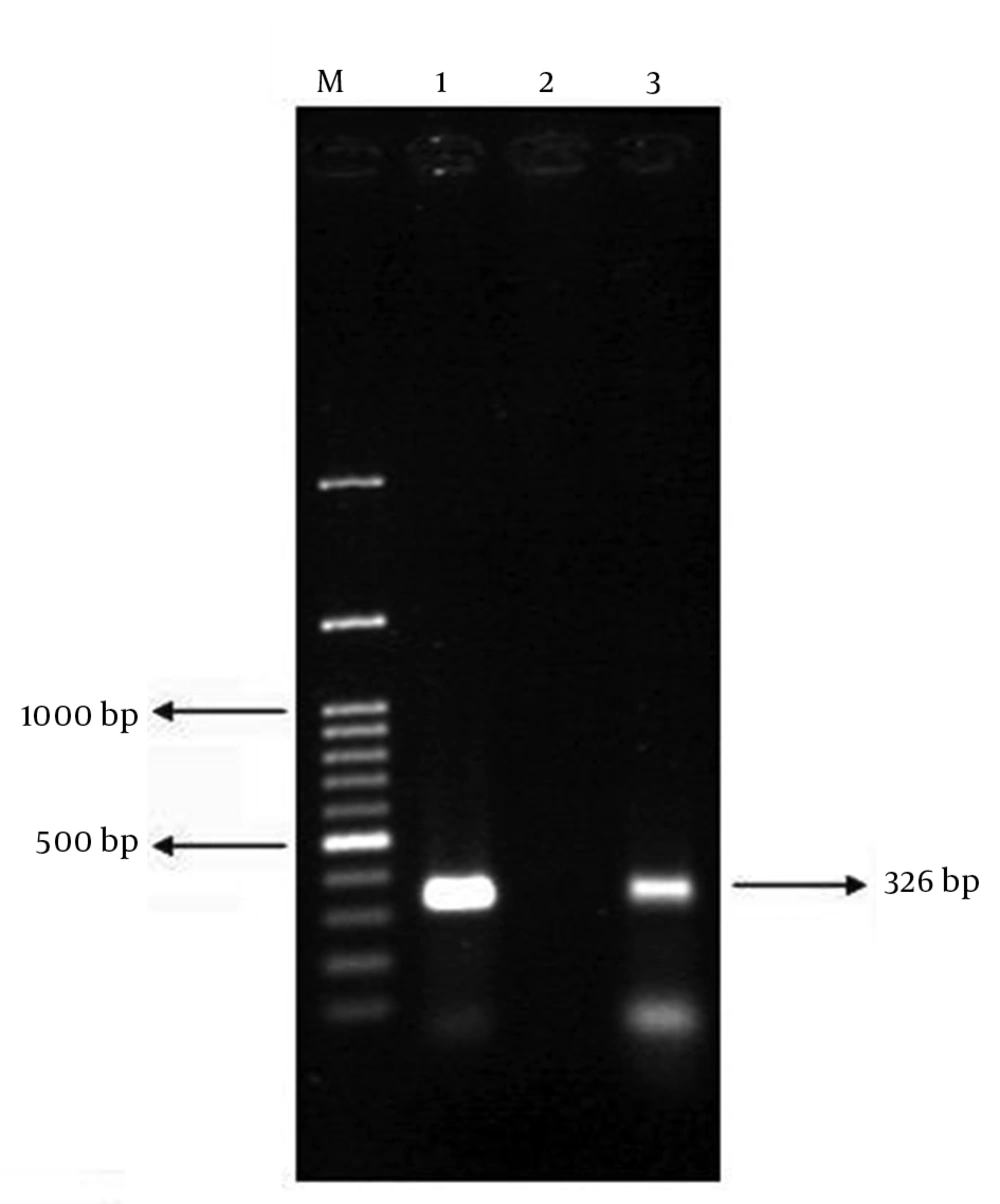
For DNA extraction, several colonies of the pure isolate were suspended in 500 μL of distilled water and heated at 100°C for 10 minutes. Then, it was centrifuged at 8000 g for 8 minutes. The supernatant was used as PCR template. To confirm EPEC strains, each E. coli isolate was examined by PCR with specific eae primers (Cinagen Co., Iran) using Eppendorf thermal cycler (Eppendorf AG, Germany) (11). Table 1 summarizes the primer sequences, annealing temperatures and the expected size of the PCR products. PCR was performed in a reaction mixture with total volume of 25 μL, containing 20 μL sterile water, 50 ng of template DNA, 2.5 μL 10X Taq polymerase buffer, 0.3 μL dNTPs (10 mmol/L), 1 U Taq DNA polymerase and 0.4 mol/L of each primer. The PCR program consisted of an initial denaturation step at 94°C for 4 minutes, followed by 32 cycles of denaturation at 93°C for 30 seconds, annealing at 55°C for 30 seconds and extension at 72°C for 40 seconds. After the last cycle, a final extension at 72°C for 4 minutes was performed. Three microliters of PCR product were analyzed by gel electrophoresis with 1% agarose (Bio-Rad, United States). Gels were stained with ethidium bromide and visualized by UV transillumination. The presence of stx1, stx2 and bfp genes in eae-positive strains was further evaluated by PCR (12, 13). Reactions were performed in the same conditions as described above, except for annealing temperatures (Table 1). E. coli strains ATCC 2348/9 (eae and bfp) and EDL933 (stx1 and stx2) were used as positive controls (9). The isolates were preserved in tryptic soy broth with 20% glycerol and stored at -70°C.
| Gene | Primer Sequence (5' to 3') | Size of Amplicon, bp | Annealing Temperature, °C |
|---|---|---|---|
| eae | 229 | 55 | |
| F-CTGAACCAGATCGTAACGGC | |||
| R-TGATAAGCTGCAGTCGAATCC | |||
| bfp | 326 | 58 | |
| F-AATGGTGCTTGCGCTTGCTGC | |||
| R-GCCGCTTTATCCAACCTGGTA | |||
| stx1 | 302 | 55 | |
| F-CGCTGAATGTCATTCGCTCTGC | |||
| R-CGTGGTATAGCTACTGTCACC | |||
| stx2 | 516 | 56 | |
| F-CCTCGGTATCCTATTCCCGG | |||
| R-CTGCTGTGACAGTGACAAAACGC | |||
| blaCTX-M1 | 863 | 54 | |
| F-GGTTAAAAAATCACTGCGTC | |||
| R-TTGGTGACGATTTTAGCCGC | |||
| blaCTX-M2 | 865 | 54 | |
| F-ATGATGACTCAGAGCATTCG | |||
| R-TGGGTTACGATTTTCGCCGC | |||
| blaCTX-M9 | 869 | 54 | |
| F-ATGGTGACAAAGAGAGTGCA | |||
| R-CCCTTCGGCGATGATTCTC | |||
| blaCTX-M15 | 995 | 55 | |
| F-CACACGTGGAATTTAGGGACT | |||
| R-GCCGTCTAAGGCGATAAACA | |||
| SHV | 230 | 56 | |
| F-AAGATCCACTATCGCCAGCAG | |||
| R-ATTCAGTTCCGTTTCCCAGCGG | |||
| blaTEM | 856 | 53 | |
| F-ATGAGTATTCAACATTTCCGC | |||
| R-CAATGCTTAATCAGTGAGG |
3.2. Serogrouping
Serogrouping using polyvalent antisera (Mast, United Kingdom) was performed for confirmed EPEC strains according to manufacturer's instructions. The polyvalent antisera consist of three separated pools, able to react with the following serogroups: poly group 2 (O26, O55, O111, O119 and O126), poly group 3 (O86, O114, O125, O127 and O128) and poly group 4 (O44, O112, O124 and O142). Strains agglutinated with polyvalent antisera were retested by monovalent O antisera.
3.3. Antibiotic Susceptibility Testing
The antibiotic susceptibility testing was performed on Mueller-Hinton agar by disk diffusion method. The following antibiotics (Mast, United Kingdom) were tested as recommended by CLSI 2010 guidelines: Ampicillin (10 µg), augmentin (30 µg), ceftazidime (30 µg), cefotaxime (30 µg), trimethoprim-sulfamethoxazole (25 µg), ciprofloxacin (5 µg), tetracycline (30 µg), chloramphenicol (30 µg), aztreonam (30 µg), imipenem (10 µg) and cefoxitin (30 µg). E. coli ATCC 25922 was used as the quality control (14).
3.4. Phenotypic Detection of ESBL Production
The EPEC isolates with resistance to any of tested third-generation cephalosporins (i.e. ceftazidime and cefotaxime) were further analyzed using both ceftazidime (30 µg)/ceftazidime (30 µg) combined with clavulanic acid (10 µg) and cefotaxime (30 µg)/cefotaxime (30 µg) combined with clavulanic acid (10 µg). CLSI suggests making disks by adding 10 µL of a 1000 µg/mL stock solution of clavulanic acid to cefotaxime and ceftazidime disks. ESBL production was confirmed if the zones produced by the disks with clavulanate were ≥ 5 mm larger than those without the inhibitor (15).
3.5. Molecular Detection of β-Lactamase Genes
PCR was performed for detection of blaCTX-M1, blaCTX-M2 and blaCTX-M9 genes (16). The isolates positive for blaCTX-M1 group were further analyzed by PCR with blaCTX-M15 specific primers (17). All isolates were also screened for blaSHV and blaTEM genes (Table 1) (18, 19). PCR conditions were the same as described above, except for annealing temperatures (Table 1). Clinical strains carrying those β-lactamase genes were used as positive controls.
3.6. Determination of Minimum Inhibitory Concentrations (MICs)
All isolates harboring blaCTX-M were subjected to MIC testing for ceftazidime and cefotaxime by agar dilution according to CLSI 2010 guidelines. E. coli ATCC 25922 was used as control strain (14).
3.7. Genotyping of CTX-M15-Producing EPEC Strains by MLVA Analysis
Multi Locus VNTR Analysis (MLVA) was performed for isolates with positive results for blaCTX-M15 gene as described by Gorge et al. (20). The PCR primers and repeat sizes for each locus are listed in Table 2. PCR was performed separately for each locus in a reaction mixture with total volume of 25 μL, containing 20 μL sterile water, 50 ng of template DNA, 2.5 μL 10X Taq polymerase buffer, 0.3 μL dNTPs (10 mmol/L), 1 U Taq DNA polymerase and 0.4 mol/L of each primer. The PCR program consisted of an initial denaturation step at 94°C for 4 minutes, followed by 32 cycles of denaturation at 93°C for 30 seconds, annealing at 55°C for 30 seconds and extension at 72°C for 60 seconds. After the last cycle, a final extension at 72°C for 4 minutes was performed. The size of each locus (PCR amplicon) was easily determined by gel electrophoresis with 2% agarose. The repeat copy numbers deduced for each isolate using the formula: Number of repeats (bp) = [size of each locus (bp)-flanking regions (bp)]/repeat size (bp). The null allele was given when the locus failed to amplify in PCR assay. After entering data into Microsoft Excel 2010 (Redmond, WA, USA), the distance matrix was calculated with the categorical coefficient using the web-based MIRU-VNTRplus (www.mlvaplus.net) (21). This distance matrix was imported into MEGA v5 to generate an unweighted-pair group method using average linkages (UPGMA) dendrogram (22).
| Loci | Primer Sequence (5' to 3') | Repeat Sizes at Each Locus, bp | Annealing Temperature, °C |
|---|---|---|---|
| ms06 | 39 | 55 | |
| F-AAACGGGAGAGCCGGTTATT | |||
| R-TGTTGGTACAACGGCTCCTG | |||
| ms07 | 39 | 55 | |
| F-GTCAGTTCGCCCAGACACAG | |||
| R-CGGTGTCAGCAAATCCAGAG | |||
| ms09 | 179 | 55 | |
| F-GTGCCATCGGGCAAAATTAG | |||
| R-CCGATAAGGGAGCAGGCTAGT | |||
| ms11 | 96 | 55 | |
| F-GAAACAGGCCCAGGCTACAC | |||
| R-CTGGCGCTGGTTATGGGTAT | |||
| ms21 | 141 | 55 | |
| F-GCTGATGGCGAAGGAGAAGA | |||
| R-GGGAGTATGCGGTCAAAAGC | |||
| ms23 | 375 | 55 | |
| F-GCTCCGCTGATTGACTCCTT | |||
| R-CGGTTGCTCGACCACTAACA | |||
| ms32 | 101 | 55 | |
| F-GAGATTGCCGAAGTGTTGC | |||
| R-AACTGGCGGCGTTTATCAAG |
3.8. Statistical Analysis
Fisher’s exact test (SPSS version 17.0) was used to examine the significance of association between different β-lactamase genes and the presence of bfp gene. The test is recommended for the analysis of contingency tables when sample sizes are small. The level of statistical significance was set at P < 0.05.
4. Results
Of 349 stool specimens (one stool per patient), 398 E. coli isolates were recovered. Of these isolates, 42 were EPEC. Therefore, EPEC was recovered from 12% of patients. These isolates comprised 16 tEPEC and 26 aEPEC (Figure 1). The mean age of patients was 2.7 ± 2.3 SD; 24 (57.1%) were females and 18 (42.9%) males, with a female-to-male ratio of 1.3: 1.
4.1. Serogrouping
The most common serogroups among EPEC isolates were the members of O127 (n = 5, 12%) and O142 (n = 5, 12%), followed by O86 (n = 4, 9.5%), O128 (n = 2, 4.8%), O111 (n = 1, 2.4%) and O44 (n = 1, 2.4%). However, most isolates (n = 24, 57.1%) were nontypeable (ONT) with polyvalent antisera used.
4.2. Antibiotic Susceptibilities of EPEC Strains
All isolates (n = 42, 100%) were susceptible to imipenem and cefoxitin. High resistance rates were observed against ampicillin (n = 26, 61.9%), followed by co-trimoxazole (n = 23, 54.8%), augmentin (n = 18, 42.9%), tetracycline (n = 16, 38.1%), cefotaxime (n = 9, 21.4%), ceftazidime (n = 8, 19%), aztreonam (n = 8, 19%) and ciprofloxacin (n = 7, 16.7%). By contrast, only one isolate was resistant to chloramphenicol.
4.3. Phenotypic ESBL Detection
Phenotypic confirmatory test using cefotaxime in combination with clavulanic acid as an inhibitor revealed that of 42 EPEC, nine (21.4%) isolates were ESBL producers. However, only eight isolates had positive results when the test was performed with ceftazidime. An increase of 5 mm in the zone of inhibition for either ceftazidime or cefotaxime tested in combination with clavulanic acid versus its zone when tested alone indicated that the isolate was an ESBL producer.
4.4. Detection of β-lactamase Genes
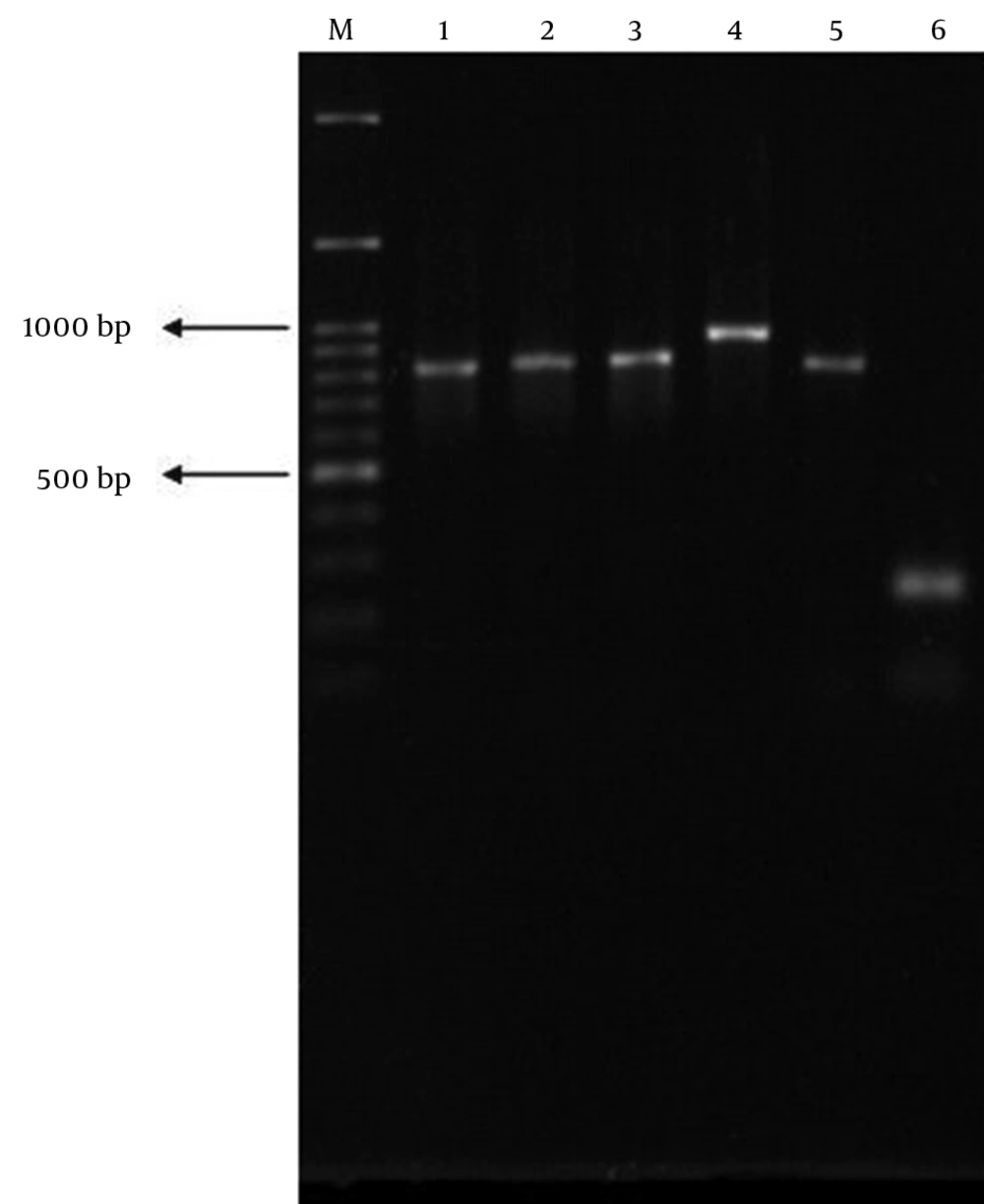
PCR was performed for all EPEC strains. Of nine ESBL producers, eight carried the blaCTX-M1. None of the isolates carried blaCTX-M2 and blaCTX-M9. Subsequent PCR revealed that all of the blaCTX-M1-positive isolates harbored blaCTX-M15 subtype. Of these eight isolates, three were tEPEC (BFP-positive), while the remaining five isolates belonged to aEPEC (BFP-negative). Statistical analysis showed that the presence of blaCTX-M15 was not associated with existence of bfp gene (P > 0.05 by Fisher's exact test). Of eight blaCTX-M15-positive isolates, only one isolate was typeable with polyvalent antisera used (Table 3). On the other hand, blaSHV and blaTEM genes were detected in 40.5% (n = 17) and 19% (n = 8) of all EPEC isolates, respectively (Figure 2). Similarly, a significant association was not observed between existence of bfp gene and presence of blaSHV or blaTEM (P > 0.05 by Fisher’s exact test). Furthermore, coexistence of blaCTX-M15 and other β-lactamase genes in a single isolate was common. The blaCTX-M15 was found to coexist with blaSHV in four isolates. One isolate harbored both blaCTX-M15 and blaTEM genes together, while the other one simultaneously carried blaCTX-M15, blaTEM and blaSHVgenes.
4.5. MIC Results
Of eight CTX-M15-positive isolates, seven isolates had MICs of 128 (μg/mL) against ceftazidime. However, all CTX-M15-positive isolates typically exhibited higher MICs (256 μg/mL) against cefotaxime (Table 3).
| Strains | bfp | Serogroups | Patient sex | Patient age, y | β-lactamase Profile | MICs, μg/ml | Resistance Profile | |||
|---|---|---|---|---|---|---|---|---|---|---|
| CTX-M15 | SHV | TEM | CTX | CAZ | ||||||
| TMU49 | - | ONT | F | 1 | + | - | - | 256 | 128 | AmpAugAtmTsCip |
| TMU106 | - | O127 | F | 1 | + | - | - | 256 | 128 | AmpAtmCip |
| TMU157 | + | ONT | F | 1.5 | + | - | + | 256 | 128 | AmpAugAtmCip |
| TMU164 | + | ONT | M | < 1 | + | + | - | 256 | 64 | AmpAugAtm |
| TMU251 | - | ONT | F | 1 | + | + | + | 256 | 128 | AmpAugCip |
| TMU329 | - | ONT | F | 4 | + | + | - | 256 | 128 | AmpAugAtmTsTe |
| TMU369 | - | ONT | M | 6 | + | + | - | 256 | 128 | AmpAtmTsTeCip |
| TMU391 | + | ONT | F | 2 | + | + | - | 256 | 128 | AmpAtmTsTeCip |
a Abbreviations: Amp, ampicillin; Atm, aztreonam; Aug, augmentin; CAZ, ceftazidime; Cip, ciprofloxacin; CTX, cefotaxime; F, female; M, male; ONT, O not typeable; Te, tetracycline; Ts, co-trimoxazole.
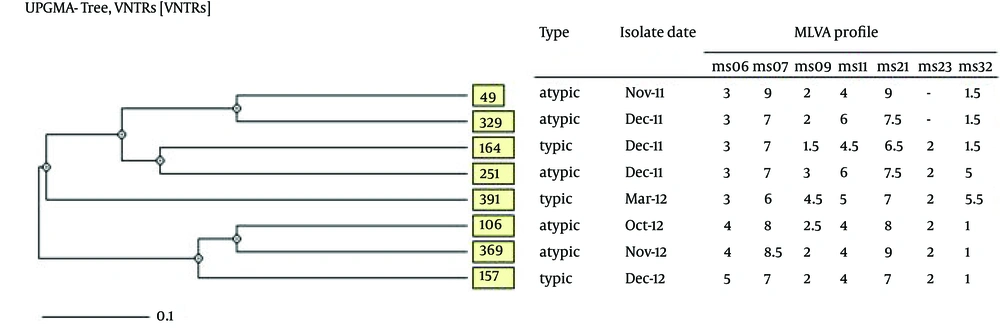
4.6. MLVA Assay
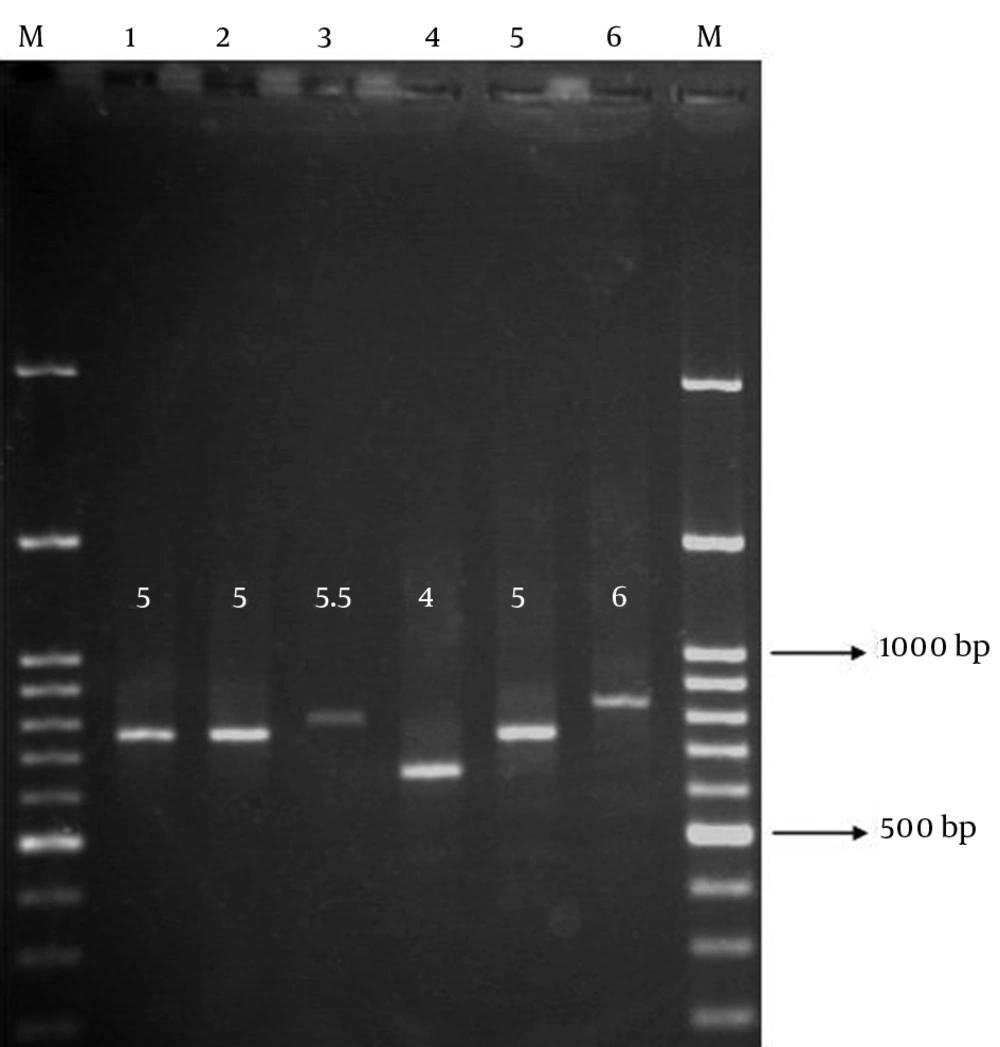
UPGMA cluster analysis revealed high genetic diversity among CTX-M15-producing isolates, which indicated lack of any significant genetic relatedness. MLVA distinguished eight unique genotypes, with the similarities ranged from 60% to 80% for the most and the least similar isolates, respectively (Figure 3). MLVA assay set-up for ms11 locus is shown in Figure 4. The image illustrates how the repeat copy numbers can be directly deduced by manual reading. The numbers above amplicons provide the repeat copy numbers for each strain.
5. Discussion
Over the last decade, blaCTX-M has become increasingly common worldwide, to the point that their prevalence easily surpassing those of blaSHV and blaTEM ESBL genes. Based on CTX-M amino acid sequences, these enzymes have been classified into five major groups, groups 1, 2, 8, 9 and 25/26. The incidence of these blaCTX-M genotypes varies geographically. The most widely disseminated genotype, the blaCTX-M15 has spread to all continents. It was first described in E. coli isolated from India during 2001. The blaCTX-M15 is sometimes associated with other genes, such as blaSHV and blaTEM as well as genes encoding for resistance to other antibiotics, such as the qnr genes conferring resistance to fluoroquinolones, and aac (6’)-Ib-cr, conferring resistance to aminoglycosides and fluoroquinolones (23).
In our country, previous studies revealed that the blaCTX-M1 was the dominant ESBL gene among clinical isolates of Klebsiella pneumoniae (24), Salmonella enterica (25) and Shigella spp. (26). Likewise, data on emergence of CTX-M15-producing Enterobacteriaceae from neighboring countries such as Kuwait (27), Saudi Arabia (28) and Turkey (29) is in accordance with our results. However, there are few reports regarding the incidence of blaCTX-M15 among diarrheagenic E. coli. In a study conducted by Albert et al. (30), the blaCTX-M28was the most common blaCTX-M variant in enteroaggregative E. coli (EAEC) and EPEC in Kuwait. Both blaCTX-M28 and blaCTX-M15 genes are members of blaCTX-M1 group. In UAE, the blaCTX-M15 was the sole variant in EAEC (31). By contrast, EPEC strains carrying blaPER-2 and blaTEM-116 ESBL genes have been recovered from children with diarrhea in South America (32). According to the study of Amaya et al. (33), EAEC was significantly more resistant than EPEC to various antibiotics in Nicaragua. They also showed that CTX-M15 enzyme was the main type of ESBL among EAEC and non-diarrheagenic E. coli isolates obtained from children with or without diarrhea (33).
Some of ESBLs evolved from older, broad-spectrum β-lactamases (e.g. blaSHV-1 and blaTEM-1). The non-ESBL SHV-1 β-lactamase has been commonly encountered in E. coli and K. pneumoniae. Additionally, the gene is usually plasmid mediated in E. coli, but chromosomally encoded in most K. pneumoniae isolates (34). In this research, almost a half of the isolates harbored blaSHV gene. Interestingly, the gene was coexisted with blaCTX-M15 in five isolates. However, most isolates expressing SHV enzyme were non-ESBL producers. Similarly, only two isolates harbored both blaCTX-M15 and blaTEM genes. The remaining TEM-positive isolates were non-ESBL producers. This finding is not surprising because TEM-1 is the most frequently encountered β-lactamase among ampicillin resistant E. coli. Although these classical β-lactamases are not considered ESBLs, their clinical importance relies on their potential to undergo mutations to increase their activity against extended-spectrum β-lactams (e.g. third generation cephalosporins) (23).
In this study, all isolates were susceptible to cefoxitin, indicating that AmpC β-lactamases did not exist among them. Cefoxitin hydrolysis as a screening marker distinguishes AmpC from other β-lactamases. These enzymes represent a clinical threat since they are not affected by β-lactamases inhibitors (e.g. clavulanic acid). Furthermore, co-existence of AmpC β-lactamases and ESBLs in the same isolate can result in false-negative phenotypic confirmatory test, because clavulanic acid induces high level expression of AmpC β-lactamases (35). As for imipenem, none of the isolates were resistant to it, indicating that carbapenemases, especially those originated from molecular class A (e.g. KPC) were absent among our EPEC isolates (36). The most predominant O-serogroups were O127, O142, O86 and O128 in our study. Interestingly, most of CTX-M15-positive isolates were not typeable with diagnostic antisera. In a study conducted by Alikhani et al. (9) serogroups O127, O142, O111, O55 and O26 were the major groups, which were somewhat different from those of our work. On the other hand, Blanco et al. (37) showed that O55, O111 and O119 were the most common serogroups in Uruguay, whereas another study from Germany reported that O26, O55, O86 and O128 were the most prevalent serogroups (38). Serogroups can also vary over time, by region, or even by same regions inside a country.
In our research, most isolates were not typeable with the panel of diagnostic antisera. Indeed, the diversity of serogroups among EPEC strains discouraged the use of serotyping methods for their diagnosis (37-39). MLVA is a promising genotyping method, which is easy to perform, cheap and highly reproducible alternative to Pulse Field Gel Electrophoresis (PFGE) and Multi Locus Sequence Typing (MLST). MLVA has been used successfully to genotype ESBL-producing E. coli strains in Denmark (40). It was also applied for genotyping E. coli strains carrying different groups of CTX-M family in some European countries (41).
The most obvious finding from this survey was that there was a high degree of heterogeneity among ESBL producing strains. The presence of these strains was not due to emergence of one specific clone, but seemed to be due to the spread of mobile genetic elements (e.g. plasmids and transposons) harboring blaCTX-M15 as well as other β-lactamase resistance genes. In conclusion, this study emphasized the alarming role of β-lactamases, especially ESBLs in antibiotic resistance in diarrheagenic E. coli strains. It gave us an insight into the current prevalence and genetic backgrounds of these strains.