1. Background
Acinetobacter baumannii infections take place particularly in patients with serious underlying disorders, immunosuppressed ones, in patients undergoing invasive procedures, and the ones who have been treated with broad-spectrum antibiotics. Therefore, infections due to A. baumannii are commonly detected at intensive care units (ICUs) (1). Acinetobacter species are opportunistic pathogens which are prevalent in soil, waste water and hospital environment. Their colonization increases as the duration of hospitalization increases. Acinetobacter strains may colonize for approximately five months on hospital equipment (2, 3).
There was no resistance problem with Acinetobacter species in 1970s due to the limitation in broad-spectrum antibiotics usage and these isolates were not as important as today. However, recently, the wide and irrational use of broad-spectrum antibiotic has caused the emergence of multidrug resistant (MDR) pathogens in hospital settings (4, 5). Treatment of infections due to Acinetobacter species is a challenge due to relapses. More than 40 resistance genes are reported which can show genetic variance. Therefore, the strain can gain resistance to various antibiotics (6). MDR A. baumannii strains are the first line causes of infection, particularly in ICUs (7). Recently, there have been reports on MDR Acinetobacter species including aminoglycosides, quinolones, and carbapenems (2, 3, 8).
There is rapid improvement in molecular diagnostic methods for detection of infectious pathogens. This has led to detailed researches on identification, clonal relationship, antibiotic resistance, and epidemiology of hospital infections. This also helps preventing the spread of resistant strains (9). There are studies using molecular methods which were able to differentiate MDR A. baumannii strains at the clonal level. This helps controlling these resistant strains and prevents their epidemy (10-12). There are various methods in detecting the epidemiology of hospital-acquired A. baumannii isolates.
2. Objectives
The aim of our study was to investigate the antimicrobial susceptibility and clonal relationship of A. baumannii strains isolated from wound, blood and tracheal secretion specimens by enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC PCR) at the ICU of a university hospital in Turkey.
3. Materials and Methods
3.1. Bacterial Isolates, Identification and Antibacterial Susceptibility
Thirty three A. baumannii strains isolated from wound, blood, and tracheal secretion specimens between January 1st 2013 and June 30th 2013 at the Izmir University School of Medicine Hospital ICU were included. The clinical specimens were cultivated on blood agar, chocolate agar, and eosin methylene blue (EMB) agar (Salubris, Turkey). The plates were incubated at 37оC for 48-72 hours. One isolate per patient was used in the study. The identification and antimicrobial susceptibility of the strains were studied by automatized Vitek version 2.0 (Biomerieux, France). The Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853 and Klebsiella pneumoniae 700603 were used as quality control strains.
3.2. Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction
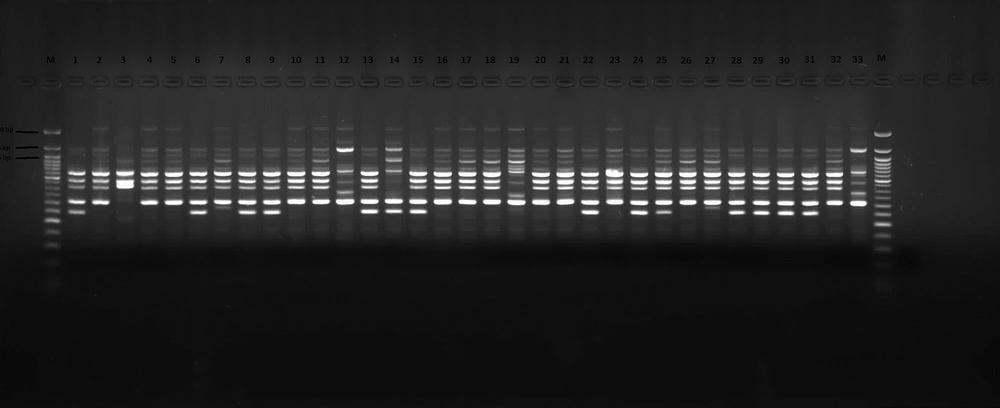
The clonal relationship among the A. baumannii strains was analyzed using ERIC-PCR (5’-AAG TAA GTG ACT GGG GTG AGC G-3’) (Fermentas, Lithuania) (13). To evaluate similarity between the isolates, Jaccard coefficients were derived from the banding patterns (14). Dendrograms were constructed according to the unweighted pair group method with arithmetic mean method, using Jaccard coefficients and MEGA software version 4.0 (15). The components of the 50 µL master mix are shown in Table 1. The amplification was as follows: one cycle (94ºC for one minute), 30 cycles (94ºC for one minute, 45ºC for one minute, 72ºC for two minutes), and 72ºC for five minutes. The amplified products were imaged using a 0.5 µg/mL ethidium bromide-containing gel, prepared in 1% agarose in 1x Tris-borate-EDTA (TBE) buffer (Sigma, USA) under 120 v for 45 minutes and under the UV light. Gene RulerTM 50 bp DNA Ladder (Biolabs, New England) was used in detecting the size of the bands.
| Components of the Master Mix | Data |
|---|---|
| 10x Enzyme Buffer, | 5 µL (1x) |
| MgCl2 | 5 µL (2.5 mM) |
| DNTPs | 5 µL (200 µM) |
| ERIC-2 | 1 µL (25 pmol) |
| Taq DNA polymerase (Fermentas, Lithuania), | 0.5 µL (2.5 U) |
| Sterile deionized water | 28.5 µL |
| DNA | 5 µL |
Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction Master Mix a
4. Results
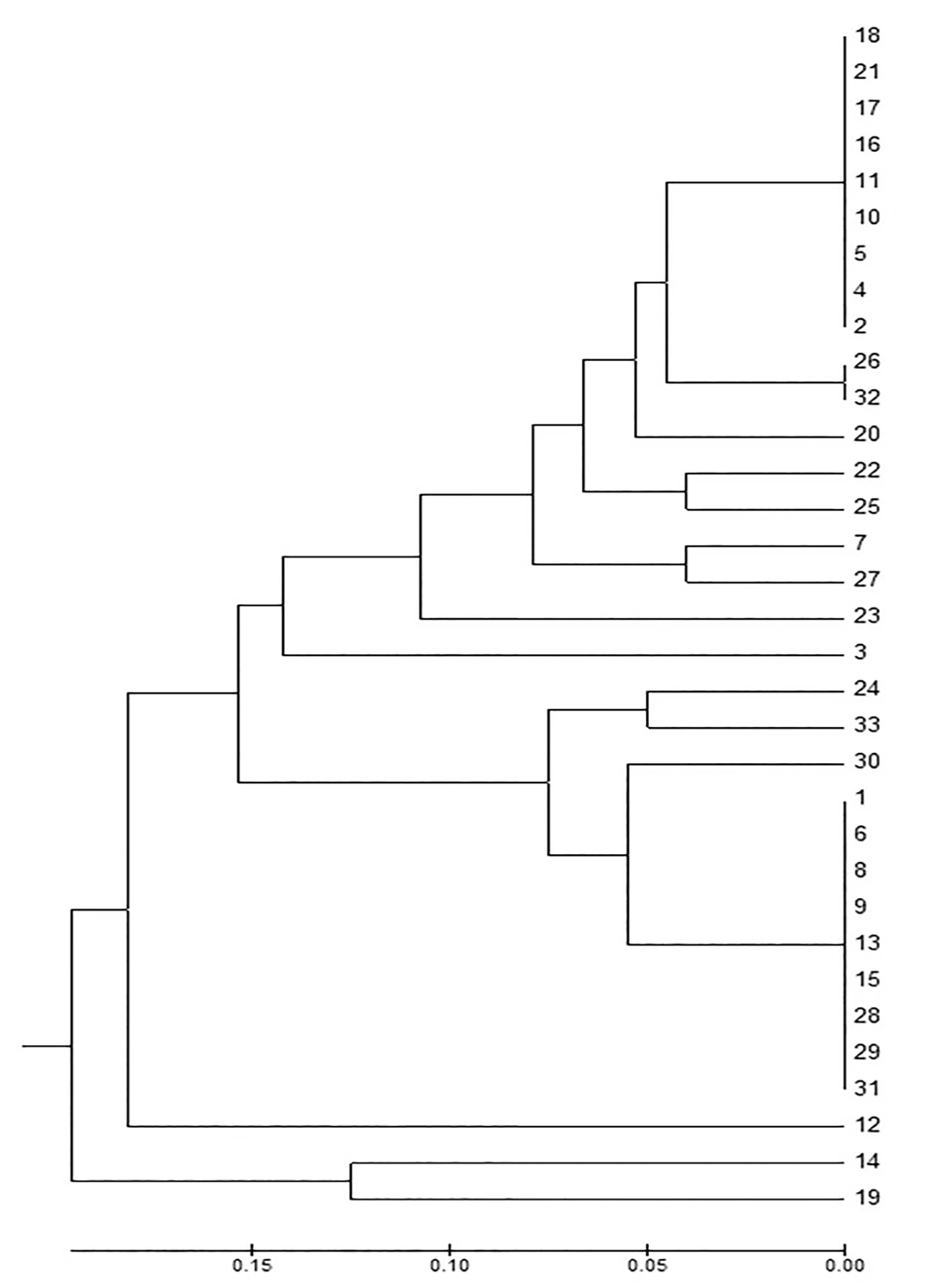
A total of 33 A. baumannii strains were included in this study. The samples consisted of tracheal secrete (23), wound (2) and blood cultures (8). The antimicrobial susceptibility of A. baumannii isolates are shown in Table 2. The ERIC-PCR gel image of the isolates is shown in Figure 1. A. baumannii complex strains were classified into seven clusters based on the fingerprinting results. The larger clusters consisted of 16 and 12 members and other clusters included only one member each (Figure 2). Our results revealed that two main clusters were responsible for the prevalence of A. baumannii complex strains at the ICU.
| A. baumannii | CAZ | FEP | TZP | IMP | MEM | AK | GN | TG | CT | SCF |
|---|---|---|---|---|---|---|---|---|---|---|
| Tracheal secretion (n = 23) | 0 | 0 | 0 | 2 (8.6) | 1 (4.3) | 9 (39.1) | 5 (21.7) | 18 (78.2) | 23 (100) | 0 |
| Blood (n = 8) | 1 (12.5) | 1 (12.5) | 1 (12.5) | 1 (12.5) | 1 (12.5) | 4 (50) | 3 (37.5) | 8 (100) | 8 (100) | 2 (25) |
| Wound (n = 2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (50) | 2 (100) | 0 |
5. Discussion
A. baumannii is a healthcare-associated pathogen, increasingly being reported as the cause of nosocomial infections, especially at ICUs (16-18). Infection with MDR A. baumannii strains has a longer duration at ICUs and hospitals. Besides, the mortality rates and treatment costs are higher among these patients (19, 20). Surveillance systems for nosocomial infections must be rapid, reliable, and capable of distinguishing between related and unrelated bacterial strains, particularly in ICUs (21, 22).
There have been studies using molecular methods which have been able to differentiate MDR A. baumannii strains at the clonal level. This helps controlling these resistant strains and prevents their epidemy (10-12). Microbial typing methods have become an essential section of clinical microbiology laboratories. These methods consist of PCR multilocus enzyme electrophoresis (MLEE), multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), restriction fragment length polymorphisms (RFLP), DNA sequencing, ribotyping, randomly amplified polymorphism DNA (RAPD), amplified fragment length polymorphism (AFLP) and repetitive sequence-based PCR (REP-PCR) (23). The REP-PCR method uses primers that target noncoding repetitive sequences interspersed throughout the bacterial DNA and is an established approach for subspecies classification (24). The method is easy to apply and has the advantage of working with many isolates at the same time. Besides, due to its high resolution, phylogenetic analysis can be carried out. There are also disadvantages in ERIC; primers being affected by PCR conditions and band profiles may not be distinguishable (25).
Mathai et al. (26) worked with random amplified polymorphic DNA (RAPD) method to detect the molecular epidemiology of 27 hospital-acquired A. baumannii strains isolated from respiratory tract specimens. The authors have detected various clones at the hospital. In our study, The ERIC-PCR gel image of A. baumannii complex strains indicated seven clusters based on the fingerprinting results. Yong et al. (27) reported 23 different patterns among PER-1-positive Acinetobacter strains by PFGE method. Most of the strains were isolated from ICU which may indicate clonal spread. Our study indicated seven clusters. In another study in Turkey; the clonal relationship between nosocomial A. baumannii strains was detected by RAPD-PCR and PFGE; 80% of 41 A. baumannii strains belonged to genotype 3. Thirty two patients were hospitalized in ICU and thus, a clonal spread was considered (28). In our study, the largesr clusters consisted of 16 and 12 members and other clusters included only one member each.
Akalin et al. (29) detected 12 different genotypes in 120 A. baumannii isolates during a three-year period by RAPD-PCR method. The strains consisted of surveillance isolates, clinical strains and ICU isolates. The authors considered this data as cross-contamination and environmental contamination due to A. baumannii.
Carbapenems are the first line therapy for MDR A. baumannii strains. Increase in carbapenemase production resistance has been detected due to its over usage. Ozdem et al. (30) reported carbapenem resistance 32% in 2007, which increased to 80% in 2010 among A. baumannii strains. Irrational and prevalent use of antibiotics increases the resistance rate and causes trouble in treatment. In our study, carbapenem resistance was high similar to other studies. The resistance rate was 91.4% in tracheal secretion samples, 87.5% in blood cultures and 100% in wound cultures. Colistin is an effective antibiotic for MDR A. baumannii isolates (31). Dizbay et al. (32) reported 100% susceptibility to colistin for A. baumannii isolates. In our study, we also detected 100% susceptibility to colistin for A. baumannii strains. Besides, tigecycline was the second most susceptible antimicrobial agent.
MDR A. baumannii strains cause trouble in treatment. Increased resistance has led to an increment in morbidity, mortality, and cost. Therefore, molecular studies on clonal relationship and data on antimicrobial resistance are important in preventing epidemy and initiating effective treatment, especially in MDR strains such as A. baumannii. ICUs are sections of hospitals where invasive procedures (such as urinary catheterization and mechanical ventilation) are frequently held and a wide spectrum of antibiotics are administered. This may be the reason of detection of Acinetobacter infections at these units. The use of molecular epidemiological methods can help us with detection of the pathogen circulation at our university hospital.