1. Background
Haemophilus influenzae is a Gram negative rod-shaped bacterium (1), that is classified as either unencapsulated or encapsulated (2). Haemophilus influenzae type b includes 90% of cases half of which appears as meningitis (3). High levels of multidrug resistance are usually associated with mobile genetic elements that encode specific resistance genes (4). Antibiotic resistance can be obtained as a result of mutation or acquisition of resistance genes by means of horizontal gene transfer, or as a natural characteristic of an organism (5). Integrons are important genetic elements that are involved in spreading antibiotic multi-resistance (6, 7). Integrons are composed of three important elements necessary for the capture of exogenous genes: a gene (intI) encoding an integrase belonging to the tyrosine-recombinase family; a primary recombination site (attI) located immediately adjacent to intI; and a strong promoter (Pc) that directs transcription of the captured genes.
In special cases, large exogenous segments (10 - 200 kb) in bacterial genomes form genomic islands (8), and one of the functions of these genomic islands is antibiotic resistance (9). Genomic islands have been considered as fragments of DNA obtained via horizontal gene transfer with major characteristics including, GC content that is normally different from the rest of the genome, ordinary insertion in tRNA genes, direct repeated DNA sequences at the ends, and the presence of genes such as integrases, transposases, or insertion sequences (10). Due to geographical heterogeneity in the antibiotic resistance pattern, it is mandatory to determine resistance patterns that are region-specific rather than generalized (11).
2. Objectives
The objective of this study was to detect class 1 and 2 integrons in clinical isolates of H. influenzae.
3. Patients and Methods
3.1. Isolation and Culturing
Twenty clinical isolates were collected from different patients admitted to the Milad hospital. These twenty isolates were identified according to the standard methods of microbiology (6). Culturing was done by growth on Chocolate agar medium (Oxoid, UK). These plates were then incubated at 37°C, in a CO2 atmosphere for 24 - 48 hours. Haemophilus influenzae was identified by the presence of tiny, moist and smooth gray colonies and absence of hemolysis, as well as positive catalase and oxidase test, presence of growth factors X and V, and other biochemical characteristics (11). Brain-heart infusion (BHI) broth (Difco, USA) was used in order to ease in bacteria assemblage for DNA purification. These tubes were then incubated at 37°C in a shaking incubator for 24 - 48 hours.
3.2. Antimicrobial Susceptibility Test
Antimicrobial susceptibility test was performed using the standard disk diffusion method on Muller-Hinton agar (Merck, Germany) according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI 2009). Disks prepared by the MAST Company (Mast Co, Merseyside, UK) were used to determine the susceptibility of isolates to Amoxicillin (A: 10 µg), Ciprofloxacin (CIP: 5 µg), Ceftriaxone (CRO: 30 µg), Clindamycin (CD: 2 µg), Azithromycin (ATH: 15 µg), Chloramphenicol (C: 30 µg), Tetracycline (TE: 30 µg), Trimethoprim (TM: 5 µg) and Sulfamethoxazole (SXT: 30 µg). Haemophilus influenzae ATCC 9006 was used as the quality control strain.
3.3. DNA Purification and Polymerase Chain Reaction for Detection of Class 1 and 2 Integrons
Total DNA of all isolates was prepared by boiling methods as previously described (12). The Purification Kit (GeneJET Plasmid Miniprep Kit) was used for DNA extraction according to the methods recommended by the manufacturer (Thermo Scientific, Fermentas, USA). All isolates were screened for detection of class 1 and 2 integrons by universal primers, designed for the intI1 and intI2 genes, respectively. Amplification was performed in a total volume of 20 μL (18 μL of PCR master mix and 2 μL of extracted DNA as template) and carried out in a Thermo cycler (Eppendorf Mastercycler®, MA) using the following cycling program: initial denaturation at 94°C for four minutes and 35 cycles of one minute at 94°C, 30 seconds at 58°C, one minute at 72°C, with a final extension for ten minutes at 72°C (6).
3.4. Gradient Polymerase Chain Reaction and Detection of Genomic Islands Integrase Gene
The selection of the annealing temperature is possibly the most critical component for optimizing the specificity of a PCR reaction. In most cases, this temperature must be empirically tested. Gradient PCR was done (by Mastercycler® gradient) in order to characterize the adaptable annealing temperature.
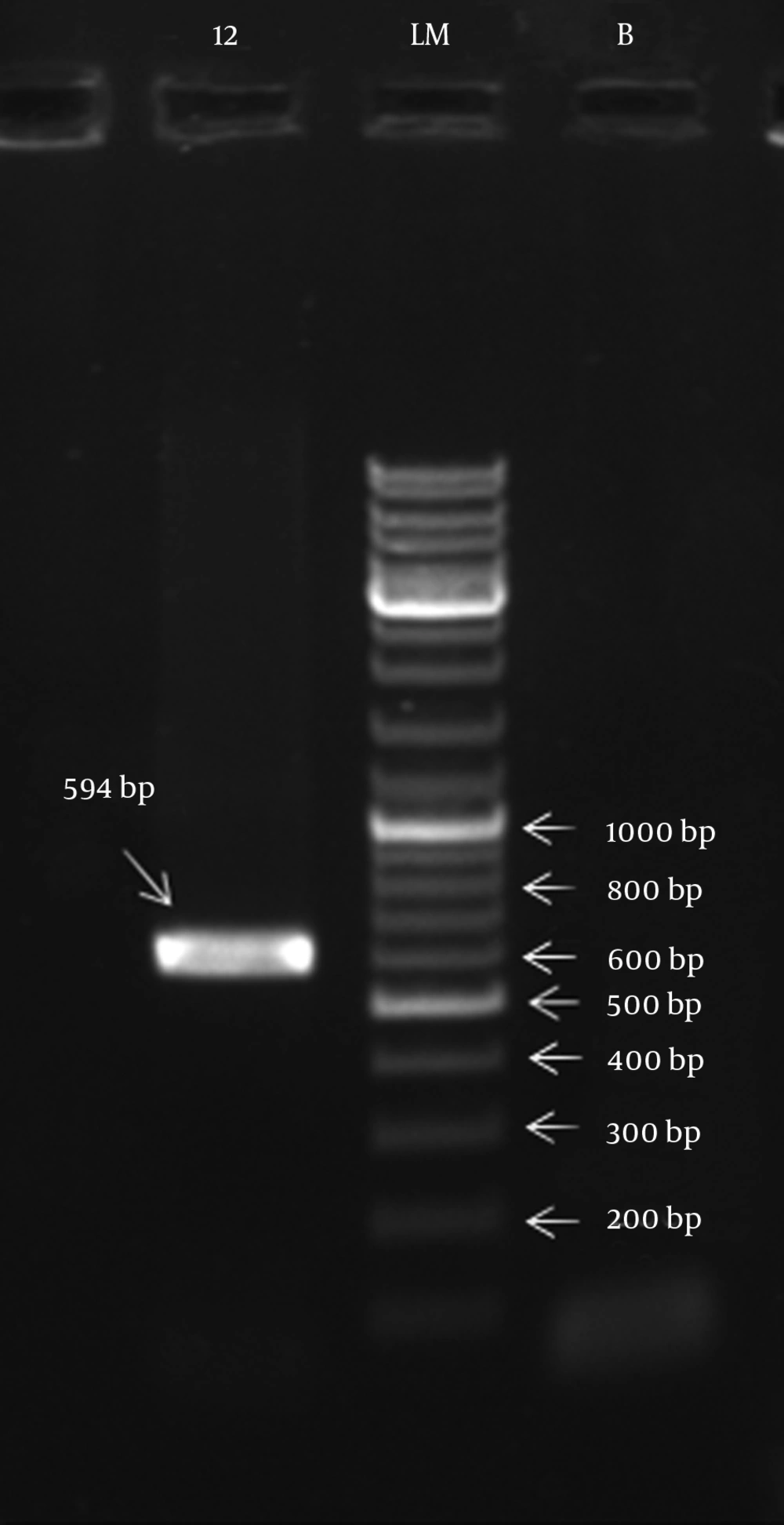
The primers used for amplification of genomic islands integrase gene were: 5’-F- GGC CTT AGG CGG TTA TCC AG-F-3’ and 5’-R- TCC GCA TTT ACC GCC TCT TT-R-3’. These primers were designed by our researchers and resulted in amplicon size of 594 bp (Phishgam Biothec Co, Iran).
The amplification program was performed by a thermocycler (Eppendorf Mastercycler®, MA) and was started with initial denaturation for four minutes at 94°C and programmed with 30 cycles of each: 45 seconds at 94°C, 45 seconds at 54°C, 45 seconds at 72°C. The program finished with the final extension of ten minutes at 72°C (13). Amplification products were extracted from the gels with the GeneJET Gel Extraction Kit (Thermo Scientific, Fermentas, USA) before sequencing.
3.5. DNA Sequencing
The PCR products of 594 bp were sequenced by the Sequencing Lab of the Pasteur Institute of Iran. The resulting DNA sequence was analyzed using the BLAST algorithm software available at http://www.ncbi.nlm.nih.gov/BLAST/. genomic island (GI) sequences were deposited in the EMBL/GenBank database (www.ncbi.nlm.nih.gov).
4. Results
4.1. Disk Diffusion
In order to assess the antibiotic resistance pattern of H. influenzae, the EXCEL software (Microsoft Office 2010) was used to analyze the antibacterial susceptibility test results. From the total of 20 H. influenzae isolates, 19 isolates (95%) were resistant to Amoxicillin, nine (45%) to Ciprofloxacin, 20 (100%) to Clindamycin, Chloramphenicol and Tetracycline, ten (50%) to Ceftriaxone and two (10%) to Azithromycin. Also 20 (100%) were sensitive to Sulfamethoxazole and 19 (95%) to trimethoprim. Multidrug resistance (MDR) is defined as being intermediate or resistant to two or more classes of antibiotics. Resistance to tetracycline, chloramphenicol and amoxicillin was found in 95% (19 of 20) of the isolates. Furthermore, 45% (9 of 20) of isolates were resistant to TE, C, A and CIP, 50% (10 of 20) to TE, C, A and CRO, 40% (8 of 20) to TE, C, A, CIP and CRO, and 5% (1 of 20) to TE, C, A, CIP, CRO and ATH, simultaneously (Tables 1 and 2).
4.2. Detection of Class 1 and 2 Integrons and Genomic Islands Integrase Gene
Of the twenty isolates, twelve (60%) isolates amplified the integrase gene of genomic islands with an amplicon size of 594 bp (Figure 1) whereas class 1 and 2 integrons were not detected in any of the isolates.
4.3. Nucleotide Sequencing of Integrase Gene Attributed to Genomic Islands
The nucleotide sequences of integrase gene related to genomic islands was deposited in the NCBI GenBank sequence databases under the accession number KC456064, available at http://www.ncbi.nlm.nih.gov/nuccore/KC456064.
| Antibiotics b | Resistance | Sensitive | Intermediate |
|---|---|---|---|
| A | 19 (95) | 1 (5) | 0 (0) |
| CIP | 9 (45) | 11 (55) | 0 (0) |
| CD | 20 (100) | 0 (0) | 0 (0) |
| C | 20 (100) | 0 (0) | 0 (0) |
| TE | 20 (100) | 0 (0) | 0 (0) |
| CRO | 10 (50) | 10 (50) | 0 (0) |
| ATH | 2 (10) | 18 (90) | 0 (0) |
| TM | 0 (0) | 19 (95) | 1 (5) |
| SXT | 0 (0) | 20 (100) | 0 (0) |
a Data are presented as No. (%).
bA, Amoxicillin: 10 µg; CIP, Ciprofloxacin: 5 µg; CRO, Ceftriaxone: 30 µg; CD, Clindamycin: 2 µg; ATH, Azithromycin: 15 µg; C, Chloramphenicol: 30 µg; TE, Tetracycline: 30 µg; TM, Trimethoprim: 5 µg; SXT, Sulfamethoxazole: 30 µg.
| Antibiotics b | TE,C,A | TE,C,A,CIP | TE,C,A,CRO | TE,C,A,CIP,CRO | TE,C,A,CIP,CRO,ATH |
|---|---|---|---|---|---|
| Resistant | 95% | 45% | 50% | 40% | 5% |
a Data are presented as No. (%).
bA, Amoxicillin: 10 µg; CIP, Ciprofloxacin: 5 µg; CRO, Ceftriaxone: 30 µg; CD, Clindamycin: 2 µg; ATH, Azithromycin: 15 µg; C, Chloramphenicol: 30 µg; TE, Tetracycline: 30 µg; TM, Trimethoprim: 5 µg; SXT, Sulfamethoxazole: 30 µg.
5. Discussion
Genetic transformability of bacteria induces the SOS response to antibiotic stress-induced DNA damage and leads to promotion of horizontal dissemination of antibiotic resistance genes (14). Organisms are protected from the effects of antimicrobial agents because of bacterial resistance to antibiotics caused by genetic variations (15). Integrons, as mobile genetic elements, give bacteria ecological advantages in the face of environmental conditions and thus have an important role in multidrug resistance dissemination (16). One of the most frequent traits associated with genomic islands is antibiotic resistance. As there are numerous studies regarding the emergence of antibiotic resistance in H. influenzae, these bacteria can be good models for how resistance genes have become associated with a genomic island.
Genomic islands have an important effect on evolution and adaptation of microorganisms such as distribution of virulence genes and antibiotic resistance, and formation of catabolic pathways (14). Genomic islands facilitate horizontal transfer of genes in bacteria, including transfer of antibiotic resistance. Resistance pattern to amoxicillin, tetracycline, chloramphenicol and erythromycin was identified in genomic islands of H. influenzae (17). Herein there is a homology between integrons and genomic islands. Overall, 95% of our studied isolates were phenotypically resistant to TE, C and A, yet Int1 and Int2 genes were not detected in any of these isolates. Therefore, genotypic evaluation was done on integrase genes attributed to genomic islands and this gene was detected in 12 of the 20 isolates. Variations between phenotypic and genotypic results are justified because of different pathways for resistance.
An integron is a two component gene capture and dissemination system, initially discovered in relation to antibiotic resistance, and is found in plasmids, chromosomes and transposons. The first component consists of a gene encoding a site-specific recombinase along with a specific site for recombination, while the second component comprises of fragments of DNA called gene cassettes, which can be incorporated or shuffled. A cassette may encode genes for antibiotic resistance (18). The gene cassettes are excised and integrated by integrase from and to the integron (19). Sometimes gene cassettes are not expressed on integrons. Also, there are isolates that have the resistance phenotype yet they don’t have its gene cassette on the integron. In these cases, other factors and genetic elements, except integrons, are responsible for antibiotic resistance (20, 21). The results of the present study confirm this point.
According to the findings of previous research, the integrons of class 1 are strongly resistant to Trimethoprim and Sulfamethoxazole (22-27). This study showed that all isolates were sensitive to Trimethoprim and Sulfamethoxazole, which can be an indicator of the absence of class 1 integrons or it can be due to the above-mentioned geographical heterogeneity, since isolates of this study were native to Iran, hence had a different genetic base. The findings of this study suggest that genomic islands can be an efficient source for gaining antibiotic resistance genes. In this sense, the findings seem to justify the presence of genomic islands in H. influenzae isolates as well as the multiple resistances of drugs in these isolates.
Treatment of some bacterial infections, such as those caused by Gram-negative bacteria, are already very difficult. If development of new resistance mechanisms is continued by bacteria, and if novel classes of antibiotics are produced by pharmaceutical industries at the existing rate, the future for medicine is bleak (28, 29). Hence, importance of antimicrobial susceptibility monitoring is clear for the detection of resistant clinical isolates and for the surveillance of antimicrobial resistance (30). Resistance to a variety of antibiotics has increased significantly in bacterial pathogens. An idea behind the increment of resistance is the ability of microorganisms to receive resistance genes from far related genera. The interaction of gene transfer elements with each other would enhance microorganism's ability to transfer resistance genes collectively. When considering the potential for horizontal transfer of resistance genes, this interactive capacity needs to also be taken into account (31). In conclusion, the crucial role of genomic islands (GIs), integrons and other genetic elements in dissemination of variable genes, including antibiotic resistance genes, is highly important for avoiding generation and distribution of multidrug resistance.