1. Context
Bacterial pathogens and their toxins cause illnesses, which spread throughout the population. Some bacteria are producing enterotoxins such as cholera toxin, the heat-labile or heat-stable enterotoxins produced by Escherichia coli. Others produce cytotoxins like shiga toxins produced by Shigella, which damage cells. Both of them can cause diarrheal diseases (1, 2). When pathogenic bacteria overcome host microbiome of normal flora, diarrhea develops (3). In 1980s, the main mortality rate of diarrhea was approximately 4.6 million per year, but it has decreased to 1.6-2.1 million since then. Most of these deaths occur in infants and young children under the age of 5 years in developing countries. Diarrhea has a lot of symptoms such as nausea, vomiting, fever, and abdominal pain. Important risk factors of diarrhea consist of age, gastric acidity, antibiotics, immunosuppression, and poor sanitation (4). One kind of this syndrome is travelers' diarrhea (TD) that is the most common cause of disability among international travelers to developing countries. Nowadays, infectious diarrhea has become one of the main health problems worldwide.
2. Evidence Acquisition
A rapid detection method, including identification of the pathogens in population is critical in the disease control (5). The major causes of TD are E. coli, Shigella spp., Campylobacter spp., Salmonella spp., Aeromonas spp., Plesiomonas spp., and non-cholera Vibrios. Since 1970s, enterotoxigenic E. coli (ETEC) has been the most important pathogen responsible for TD (6). Novel and important objectives in identification of the enteric bacteria are development of efficient, rapid, and simple methods to detect microorganisms (7, 8). We could classify rapid methods into modified conventional methods, biosensors, immunological methods, and nucleic acid based assays, which are being described in this article.
3. Results
3.1. Conventional Detection Methods
In these methods, detection of bacteria and viruses mainly depends on the culture of the food sample (using microbiological media), biochemical identification of bacterial genera, or cell culture techniques (9). These methods are sensitive and inexpensive, but they are both time- and material-consuming due to its initial enrichment (a minimum of 5-7 days are required to identify an isolated colony), which typically occur in a few samples. It can delay the proper diagnosis and treatment regime, resulting in longer hospital stays (10). Culture is named to describe the biological amplification of viable and cultivatable bacteria with manufactured growth media. Isolation of the specific bacterial species from a mixed culture, without pre-enrichment is difficult. Therefore, it is possible to use a magnetic separation assay by a magnetic separator (11). To improve conventional methods and reduce the costs, we used several modification in the preparation of samples, plating, and missing counting to provide faster and easier methods.
3.1.1. The Analytical Profile Index
The analytical Profile Index (API) system is a version of conventional method that is developed for quick identification of the Enterobacteriaceae family members and other Gram-negative bacteria. This system consists of a plastic strip with 20 small reaction tubes, containing the separated compartments. The API test system is manufactured by bioMerieux, Corp., Marcy Etoile, France. This assay is considered the “Gold standard” with an overall sensitivity of 79%. In this technique, a reaction occurs within 24 hours. This system is very useful for identifying pathogenic Yersinia isolates and has the highest sensitivity both at the genus and at the species level (12).
3.2. Immunological-Based Methods
Immunodetection has become a broadly used method for enteric bacteria because it permits for sensitive and specific detection. Immunological assay based on antibodies is a technology employed for the detection of bacterial cells, spores, viruses and toxins (13). Methods based on antigen–antibody interaction are used for the dedication of food-borne pathogens. Polyclonal and monoclonal antibodies are used in these methods. Although, the immunological detection methods are not as specific and sensitive as nucleic acid-based detection, they are faster, more powerful and have the ability to detect both contaminating organisms and their toxins that may not be expressed in the organism's genome. In this section, we will describe some of these methods (13).
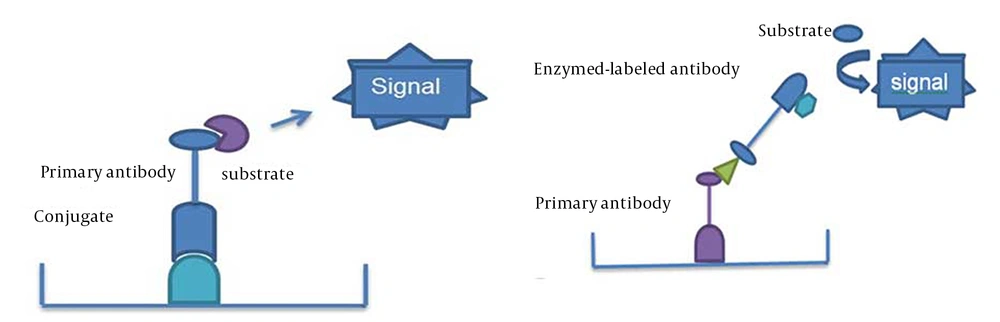
3.3. Enzyme-Linked Immunosorbent Assay
This method is only based on immunological technique and belongs to heterogeneous assays. Enzyme-linked immunosorbent assay (ELISA) binds the specificity of antibodies and the sensitivity of simple enzyme assays by using antibodies or antigens attached with an easily assayed enzyme. ELISA is an assay similar to radioimmunoassay (RIA), but using an enzyme attached with an antigen or an antibody rather than a radioactive isotope. There are several kinds of this assay such as direct ELISA, indirect ELISA, and sandwich ELISA (14).
3.3.1. Indirect Enzyme-Linked Immunosorbent Assay
In this type, the target antigen is coated in a solid phase in an ELISA plate. When serum samples are added, specific antibodies will bind the coated antigen. The ELISA plates are washed to delete unbound antibodies. Anti-immunoglobulin antisera conjugated with a peroxidase enzyme are then added. When the substrate buffer is added, in positive cases the color of the substrate buffer will change. The color is measured at a defined wavelength using a spectrophotometer, which is proportional to the level of antibodies present in the sample (14).
3.3.2. Competitive Enzyme-Linked Immunosorbent Assay
The cELISA (Competitive ELISA) can be used to detect and quantify antibody or antigen using of a competitive method. The cELISA for detection of specific antibodies has largely replaced the iELISA in large-scale screening and serosurveillance. The cELISA offers significant advantages over an iELISA, as samples from many species may be tested without the need for species-specific enzyme-labeled conjugates. Many antigens are extremely difficult or time-consuming to purify. When used in an indirect assay, they can produce high background values because of their nonspecific binding. However, relatively crude antigens may be used in the cELISA, provided that the ‘detecting antibody’ has the desired specificity. The principle of a competitive assay (for the antibodies detection) is competition between the test serum and the detecting antibody. Specific binding of the detecting antibody is detected by using an appropriate anti-species conjugate. Reduction in the obtained expected color is caused by binding of the antibodies in the test serum with the antigen, which prevents binding of the specific detecting antibody (Figure 1) (14).
3.3.3. Immunofluorescence Assay
In this way, antibodies are labeled with a fluorescent reporter molecule whose name is fluorescein isothiocyanate (FITC). This fluorescent antibody is used to directly detect bacteria in clinical specimens and applied for rapid detection of bacteria in foods (15, 16). It is to be noted that polyclonal antibodies used in the procedure lack specificity and need a well-trained microbiologist to do the test. The combination of the fluorescent antibody with DEFT were used to detect E. coli:O157:H7 in milk and apple juice (16).This assay had the sensitivity of about 103 cell/mL.
3.3.4. Immunomagnetic Separation
This method, immunomagnetic separation (IMS), utilizes paramagnetic beads (about 2-3 μm in size, about 106-108/mL), which are surface activated and can be coated with antibody by incubating in the refrigerator for varying periods of time. The unattached antibody is taken away by washing. Then, the coated beads are added to a semi-liquid mixture of food that contains antigen (toxin or whole cells in Gram-negative bacteria), thoroughly mixed, and allowed to incubate for a few minutes to several hours (for reaction of antigen with antibody-coated beads). Hence, we use this assay for isolation of biological targets from samples. It has been successful in many fields, including molecular biology, immunology, and microbiology. Cells, nucleic acids, proteins or other biomolecules can be used as magnetic targets. This method reduces time and is useful for a large number of samples (17, 18).
3.4. Molecular-Based Methods
3.4.1. Polymerase Chain Reaction
Polymerase chain reaction (PCR) was invented in 1980. This assay can detect a single copy of a target DNA sequence, and amplifies a desired region of genome into billions of copies among a complex mixture of heterogeneous sequences (19). PCR is used for the detection of the pathogenic microorganisms in food (by utilizing nucleic acid for detection). This assay has advantages over culture and other methods (for the detection of microbial pathogens) such as specificity, sensitivity, rapidity, accuracy, and capacity to detect small amounts of target nucleic acid in a sample (18, 20). PCR based methods are used for the detection of a broad range of pathogens like Staphylococcus aureus (21), Listeria monocytogenes (22), Salmonella spp.(23, 24), Bacillus cereus (24), Campylobacter jejuni (25). The different forms of PCR based on their methods are real-time PCR (25-28), multiplex PCR, and reverse transcriptase PCR (RT-PCR) (23). RT-PCR is also found as multiplex RT-PCR (29-31) and real-time RT-PCR (29, 32-34).
3.4.2. Real-Time PCR (Kinetic PCR or Quantitative real Time PCR)
In spite of the development of alternative amplification technologies, PCR stays the most used method in the research, detection, and diagnosis of pathogens. One kind of PCR method is Real-time PCR. This method provides an opportunity for rapid detection of pathogens in food (35-38). Real-time PCR combines PCR chemistry with fluorescent probe detection of the amplified product. This method is simpler to carry out compared to conventional PCR method and its test result comes much sooner too (39, 40). Two kinds of chemical agents are available for real-time PCR products: fluorescent probes that bind specifically to definite DNA sequences and fluorescent dyes that intercalate into any dsDNA (41). The simplest and most cost-effective methods employed are sequence independent DNA-binding dyes such as SYBR Green I and SYBR Gold, which bound to dsDNA (42). Therefore, sensitivity and specificity, low contamination risk, ease of performance, and speed, have made real-time PCR assay an appealing alternative to conventional culture-based or immunoassay-based testing methods (39). TaqMan PCR (Fluorescent probe based real-time PCR) amplify target nucleic acid sequences from selected microbes in the samples collected from complex biological environments (39, 42).
3.4.3. Multiplex PCR
This method (simultaneous amplification of multiple gene targets ) has been designed to use two or more primer pairs directed at pathogen-specific unique sequences within a single reaction and allows the simultaneous amplification of more than one target sequence by using multiple sets of oligonucleotides to amplify two or more targets of interest (43). This method is applied for the simultaneous detection of several foodborne pathogens. For example, simultaneous detection of E. coli O157:H7, Salmonella spp. and S. aureus. Advantages of multiplex PCR include multiple targets that are amplified significantly without extra time, cost, or sample volume; however, there have been reports that multiplexing can reduce sensitivity compared with single reactions, (because of competition). The disadvantage of multiplex PCR is the competition between oligonucleotide pairs that can reduce PCR sensitivity (44).
3.5. Microarrays
Microarray is used as large scale screening systems for simultaneous identification and is very powerful tool with greater capacity (100–1000X) compared to other molecular methods (i.e. real-time PCR) that can only analyze a small number of targets. This assay is also being used for simultaneous diagnosis and detection (45). A simple microarray includes a solid surface (such as a nylon membrane, glass slide, or silicon chips) onto that is attached small quantities of ssDNA from different known bacterial species. When ssDNA from many unknown species is exposed to these array (DNA chip), strains will bind to their individual sites on the chip.
3.6. Detection Based on Fluorescent In Situ Hybridization Assay
Fluorescent in situ hybridization (FISH) is a molecular technique, which is sensitive, rapid, and useful for many phylogenetic, ecologic, diagnostic, and environmental studies in all fields of microbiology. In this method, a specific oligonucleotide probe is labeled with a fluorochrome for simultaneous identification of the pathogens by fluorescent microscopy. FISH has some advantages over conventional cultural methods, including avoidance of inhibitory substances; identification of viable but non-cultivable cells (VBNC); rapid availability of quantitative results; simultaneous identification of different species in the same sample; relatively low cost; and easy to do. PCR is more sensitive than FISH but sensitivity of FISH detection could be increased considerably after an enrichment step. This technique is applied in food samples for the detection of foodborne pathogens such as Staphylococcus spp., E. coli, Salmonella spp., Campylobacter spp, L. monocytogenes (46).
3.7. Detection Based on Loop-Mediated Isothermal Amplification (LAMP) Assay
Traditional methods for diagnosis of the disease are carried out by culturing bacteria on agar plates followed by its phenotypic and serological properties or histological examination (47). These techniques have some disadvantages such as need for previous isolation of the pathogen and insufficient sensitivity to detect low levels of pathogen (48). Molecular techniques like polymerase chain reaction (PCR) can be used to solve those problems and increase the sensitivity and specificity of the pathogen detection (49-51). Although PCR techniques are very sensitive, need for a high-precision thermal cycler has prevented these powerful methods from being widely used in the field or by private clinics as a routine diagnostic tool. Alternate isothermal nucleic acid amplification methods such as nucleic acid-based amplification (NASBA), and loop-mediated isothermal amplification (LAMP), which require only a simple heating device, have been developed for rapid and sensitive detection of target nucleic acid (52-54).
The LAMP method can produce a tremendous amount of DNA with a few copies in less than an hour with only one type of enzyme and 4–6 different specific primers without special reagents required. One advantage of LAMP over PCR is prevention of contamination, which can occur in PCR because all steps from amplification to detection are conducted within one reaction tube under isothermal conditions. Therefore, the LAMP assay is easy and requires only a water bath or heating block to provide a constant temperature as the amplification proceeds under isothermal conditions.
3.8. Detection Based on Metagenomics Assay
Metagenomics is based on a culture-independent study on microbial populations (microbiome) by analyzing the sample’s nucleotide sequence content (55). This method amplifies and sequences the whole DNA and RNA content of a given sample, by extensive filtering of the obtained data using specific software solutions. This method is useful for random detection of existing or new pathogens. In this method, the ratio of the number of target to total amplified sequences (56), sample selection (amount of pathogens in the targeted sample), time consuming data acquisition, and data analysis time are important factors. Two limitations are currently of major concern: as the method relies on finding similarities with known pathogens, there is no solution for definition of unmatched sequences; and second, software solutions that may facilitate the interpretation of the results. To minimize these disadvantages, three solutions are currently considered, the increased pathogen load (target samples with high probability of pathogen multiplication), reducing the resulting data sets by limiting the number of targeted pathogens, and excluding the host reference sequences from data analysis and optimizing bioinformatics for “profession related” application.
3.9. Detection Based on Pulsed Field Gel Electrophoresis
Pulsed field gel electrophoresis (PFGE) is useful and a gold standard for detection of food-borne zoonotic bacteria that the most important of them are S. enterica, Campylobacter spp., E .coli, Shigella spp., Vibrio cholera, and L. monocytogenes (57, 58). This technique is based on molecular assays, culture, and isolation of the bacterial strain from the food product. By this method, we can validate a full genome; however, genes with small size such as plasmids are not visible on PFGE. Therefore microbiological culture and isolation is needed for detection before the PFGE assay (59).
3.10. Sensor-Based Pathogen Detection Systems
3.10.1. Biosensor
Biosensor technology is an analytical device converting a biological response into an electrical signal. This technology is the fastest growing method for pathogen detection compared to PCR, immunology, culture methods, and gel electrophoresis. It includes a bioreceptor element such as, a microorganism, tissue, cell, enzyme, antibody, nucleic acid, biomimic, and bacteriophage (phage), which recognizes the target analyte and a transducer base on optical, acoustical, and electrochemical signal detection, for converting the recognition event into a measurable electrical signal.
3.10.2. Phage-Based Pathogen Detection Systems
Bacteriophage is a kind of virus that infects specific strains of bacteria. Enzymes, antibodies, nucleic acids, and biomimetic materials are used as bimolecular agent detectors, which have both advantages and disadvantages. Bacteriophages are biorecognition elements for the detection of different pathogenic microorganisms. Bacteriophages (phages) are viruses that attach to specific receptors on the bacterial outside and inject their genetic material inside the bacteria. These articles have a size of 20-200 nm. They recognize the bacterial receptors by means of its tail spike proteins (e.g., the tail-spike protein of Salmonella phage P22). This recognition is highly specific. Therefore, it can be used for the typing of the bacteria and development of specific pathogen detection technologies (18, 60, 61). The recognition of antigens on the surface of bacteria by using specific antibodies is an important subject. This approach does not need any time-consuming initial preparation of the sample; nevertheless, antibodies have problems, including their costly and cumbersome preparation.
Their limited shelf life is also important in their performance. Therefore, it was demonstrated that antibodies can be substituted with bacteriophages in the bacteria detection. Phages have several advantages in this assay such as long shelf-life, stability, and easy to isolation (62, 63). The bacteriophages are used in the ELISA-based assays for detection of bacterial strains. With this assay, specific strains of S.enterica and E. coli could be detected. The sensitivity of the assay was about 105 bacterial cells/well (106/mL), which is comparable with other ELISA tests detecting intact bacterial cells without an enrichment step. The specificity of the assay depends on the kind of bacteriophage used. Bacteriophages are abundant in their environment and their preparation is simple, rapid, cheap, and easy (63).
3.10.3. Surface Plasmon Resonance
A common method, which uses reflectance spectroscopy for the pathogen detection is Surface Plasmon Resonance (SPR). Application of ELISA is rapid for screening of the samples. This assay has many advantages such as selectivity, sensitivity, easy to perform, and simultaneous detection. This method can be joined to other methods such as SPR. A biosensor is an analytical tool composed of an immobilized biological ligand that ‘feels’ the analyte, and a physical transducer, which translates this phenomenon into an electronic signal. This assay uses reflectance spectroscopy for the pathogen detection and can detect small changes. One kind of this method is SPR-based biosensors that is used for the detection of food-borne pathogens such as L.monocytogenes, Salmonella spp., E. coli O157:H7, and C. jejuni. It demonstrated the use of multi-channel SPR biosensor for the simultaneous detection of multiple target analytes from complex mixtures (64-66).
This assay can identify concentrations in the picomolar range. Biosensors in SPR are a thin metal film between two transparent media of different refractive index, for example a glass prism and sample solution. It is one of the methods that can be used for quick toxin detection. SPR allows to study interactions in real time without labeling. It has high sensitivity (the picomolar range) and a thin metal film is utilized in SPR (gold is more suitable). One molecule binds to the other one, then attached to the surface thin metal film of the gold and changes the refractive index of solutions and finally the angle of the minimum reflected intensity shifts. SPR can directly determine the bacterial and plant toxins that have large molecular weight. In comparing ELISA to SPR, it was observed that ELISA is more sensitive than SPR, but the sample treatment with ELISA lasted six hours, while with SPR the treatment duration was only 20 minutes (Table 1).
| Toxin | MW (Da) | Type of Detection | Detection Limit |
|---|---|---|---|
| Enterotoxin B | 28,400 | direct | 1.96 ng/mL |
| Enterotoxin B | 28,400 | direct | not determined |
| Enterotoxin B | 28,400 | direct | 10 ng/mL |
| Enterotoxin B | 28,400 | direct | 10 ng/mL |
| β-toxin | 35,000 | direct | not determined |
| Tetanus toxin | 150,000 | direct | 0.028 Lf/mL |
3.10.4. Detection Based on an Electrochemical Biosensor
This assay is a rapid and novel electrochemical biosensor method. In this method, polypropylene microfiber membranes are coated with a conductive polypyrrole and antibody is functionalized for the biological capture and detection of enteric bacterium. The glutaraldehyde chemical can be used for attaching to conductive microfiber membranes, till a pathogen specific antibody are covalently bound to conductive microfiber membranes and then bovine serum albumin solution is used for blocking them. In this assay, use of biosensor antibodies is useful because the benefits of the antibody-antigen reaction include high binding efficiency and specificity for detection. Advantages of antibodies have made them, especially marketable for use in food materials. Then, the membranes are exposed to pathogen cells and are washed in Butterfield’s phosphate buffer and added to a phosphate-buffer electrolyte solution. With the captured pathogen on the fiber surface, the resistance in the electro-textile electrode surface increases, which converts the biological recognition event into a measurable electrical signal, indicating a positive result. This method is generally less expensive than optical detection methods and is easier to use with turbid samples (68).
3.10.5. Detection Based on Evanescent Wave Fiber-Optic Biosensors
The development of biosensors has greatly improved the sensitivity, selectivity, and speed of the microbial pathogen and biological toxin detection. Biosensors are detection devices that use living organisms or biological molecules such as antibodies, nucleic acids, or enzymes, to recognize and bind target analytes in the sample matrix. After binding, the presence of the target analyte is detected by electrical signal, a colorimetric or fluorescent indicator reaction, or some other recognition response. Because the detection of microbial pathogens and biological toxins in food, water, and human specimens are difficult, this assay relies on immunological reactions for their capture or detection. It can identify such target analytes in minutes rather than days and directly from complex matrix samples using antibody-based assays, thus significantly improves the detection sensitivity, selectivity, and speed. In addition, live organism targets can be recovered from fiber-optic waveguides to determine microorganism viability, confirm their identification, and preserve as evidence.
This technology has the potential of rapid detection of microorganisms, toxins, and other analytes. Evanescent wave fiber-optic biosensors are biosensors that utilize evanescent wave detection techniques. Electro-magnetic waves propagate within an optical fiber by total internal reflection at the exposed surface. This process induces an evanescent electromagnetic field in any surrounding dielectric media, which decays exponentially with distance from the surface. When fluorescent probes are used with this system, bounded fluorophore molecules immediately adjacent to the fiber surface are strongly excited, and some of the fluorescent signals are coupled back into the optical fiber (69, 70).
The remaining fluorescent signals are scattered and absorbed before it can be passed through the sample. Unbound fluorophores further from the fiber surface encounter lower field strength and are not effectively excited, thereby providing considerable protection from bulk sample fluorescence. Microorganisms and toxins such as Yersinia pestis (69), E. coli lipopolysaccharide endotoxin (70), pseudexin toxin (71), Clostridium botulinum toxin A (72), Staphylococcal enterotoxin B (73), ricin (74), Bacillus anthracis, Francisella tularensis, Escherichia coli O157:H7, S. typhimurium (75), have been successfully detected with evanescent wave fiber-optic biosensors (see Table 2).
| Target | Detection limit |
|---|---|
| Bacillus anthracis, colony-forming units/mL | 105 |
| Francisella tularensis, colony-forming units/mL | 105 |
| Salmonella typhimurium, colony-forming units/mL | 105 |
| Escherichia coli O157:H7, colony-forming units/mL | 105 |
| Yersinia pestis F1 antigen, ng/mL | 50 |
| Staphylococcal enterotoxin B, pg/mL | 10 |
| Cholera toxin, ng/mL | 100 |
| E. coli lipopolysaccharide endotoxin, 10 ng/mL | |
| Ricin, ng/mL | 50 |
3.10.6. Detection Based on Rapid Bioluminescent Methods
These techniques are divided into two classes: 1) methods based on bioluminescent adenosine triphosphate (ATP) assay 2) methods based on bacterial bioluminescence. These methods are useful in the food industry and give their results in the shortest time.
3.10.7. Bioluminescent Adenosine Triphosphate (ATP) Assay
Intracellular ATP is needed for all living cells. They utilize ATP for many mechanisms during all phases of the growth. ATP is destroyed within a few minutes, therefore it can be used for detection of the microbial biomass. A rapid ATP assay based on the firefly (Photuris pyralis) was developed as a replacement for the conventional plate count methods in microbiological analysis of food. Firefly (P. pyralis) luciferase produces light with ATP and luciferin (LH2) according to this reaction: LH2 + ATP + O2 Mg++ P + AMP + PPi + CO2 + hν.
The sensitivity of commercially available manual or automated luminometers is less than 0.1 pg (around 100 bacterial cells) (77). This technique can be used for milk and milk products, meat and meat products, carbonated beverages and fruit juices.
3.10.8. Bacterial Bioluminescence
Vibrio, Photobacterium, Alteromonas, and Xenorhabdus, are the major genera of bioluminescent bacteria (those capable of emitting light). In these bacteria, the bioluminescent reaction carried out by the enzyme luciferase. This process involves the oxidation of a long-chain aldehyde and reduction of riboflavin phosphate (FMNH2), which results in the emission of blue green light.
FMNH2 + O2 + RCOH luciferase FMN + RCOOH + H2O + light (490 nm)
These properties can be used in the food industry, including the detection of specific bacterial pathogens and indicator microorganisms, spore forming organisms, lactic acid bacteria, monitoring starter culture integrity, biocide and virucide, and recovery of sublethally-injured cells. Determination of ATP by firefly luciferase is a rapid technique and is useful to detect and enumerate cells, but this assay is unable to identify bacteria. We can transfer lux genes from luminescent bacteria into specific bacteria by their bacteriophages and observe the light emissions and detect strains such as S. typhimurium, Campylobacter spp, and L. monocytogenes. The sensitivity of this method is as few as 100 cells (100 per mL). It can be used to determine Gram-positive organisms. If spores of Bacillus spp. receive lux gene, light emission is observed after germination and growth phase, which is detectable by monitoring light emission (78).
3.11. Partial List of Commercially-Available Rapid Detection Kits
The following text and tables list many of the commercially available rapid detection kites; they are classified by the principles underlying the procedure used (Tables 3, 4, 5).
| Organism | Trade Name | Assay Format | Manufacturer |
|---|---|---|---|
| Campylobacter | GENE-TRAK | probe | Neogen |
| Escherichia coli | GENE-TRAK | probe | Neogen |
| E. coli O157:H7 | Probelia | PCR | BioControl |
| Listeria | BAX; GENE-TRAKb; AccuProbe | PCR; Probe; probe | Qualicon NeogenGEN-PROBE |
| Salmonella | GENE-TRAK BAX; Probelia; BIND | Probe; PCR; PCR; phage | Neogen; Qualicon BioControl BioControl |
| Yersinia enterocolitica | GENE-TRAK | Probe | Neogen |
aAbbreviations: PCR, polymerase chain reaction; BIND, bacterial ice nucleation diagnostic.
bAdopted AOAC Official First or Final Action.
| Organism/toxin | Assay Format | Manufacturer |
|---|---|---|
| Bacillus cereus diarrhoeal toxin | ||
| TECRA; BCET | ELISA; RPLA | TECRA; Unipath |
| Campylobacter | ||
| Campyslide | LA | Becton Dickinson |
| Meritec-campy | LA | Meridian |
| MicroScreen | LA | Mercia |
| VIDAS | ELFA | bioMerieux |
| TECRA | ELISA | TECRA |
| C. perfringens enterotoxin | ||
| PET | RPLA | Unipath |
| EHEC O157:H7 | ||
| RIM | LA | REMEL |
| E. coli O157 | LA | Unipath |
| Prolex | LA | PRO-LAB |
| Ecolex O157 | LA | Orion Diagnostica |
| Wellcolex O157 | LA | Murex |
| E. coli O157 | LA | TechLab |
| O157&H7 | Sera | Difco |
| Petrifilm HEC | Ab-blot | 3M |
| Dynabeads | Ab-beads | Dynal |
| EHEC-TEK | ELISA | Organon Teknika |
| Assurance | ELISA | BioControl |
| E. coli O157 | ELISA | LMD Lab |
| Premier O157 | ELISA | Meridian |
| E. coli O157 | EIA/capture | TECRA |
| Quix Rapid O157 | Ab-ppt | Universal HealthWatch |
| VIDAS | ELFA | bioMerieux |
| Shiga toxin, Stx | ||
| VEROTEST | ELISA | MicroCarb |
| Premier EHEC | ELISA | Meridian |
| Verotox-F | RPLA | Denka Seiken |
| ETEC | ||
| Labile toxin, LT | RPLA | Oxoid |
| Stabile toxin, ST | ELISA | Oxoid |
| Salmonella | ||
| Bactigen | LA | Wampole Labs |
| Spectate | LA | Rhone-Poulenc |
| Dynabeads | Ab-beads | Dynal |
| CHECKPOINT | Ab-blot | KPL |
| 1-2 Test | Diffusion | BioControl |
| Salmonella-TEK | ELISA | Organon Teknika |
| Salmonella | ELISA | GEM Biomedical |
| Transia Plate Salmonella Gold | ELISA | Diffchamb |
| PATH-STIK | Ab-ppt | LUMAC |
| Clearview | Ab-ppt | Unipath |
| UNIQUE | Capture-EIA | TECRA |
| Shigella | ||
| Bactigen | LA | Wampole Labs |
| Enterotoxin | ||
| SET-EIA | ELISA | Toxin Technology |
| SET-RPLA | RPLA | Unipath |
| TECRA | ELISA | TECRA |
| VIDAS | ELFA | bioMerieux |
| Vibrio cholera | ||
| Cholera SMART | Ab-ppt | New Horizon |
| Cholera Screen | Agglutination | New Horizon |
| Enterotoxin | ||
| VET-RPLA | RPLA | Unipath |
aAbbreviations: ELFA, enzyme-linked fluorescent assay; ELISA, enzyme-linked immunosorbent assay; EHEC, ETEC - enterotoxigenic E. coli; LA, latex agglutination.
| Organism | Assay Format | Manufacturer |
|---|---|---|
| Coliform/E. coli | ||
| Isogrid | HGMF/MUG | QA Labs |
| Petrifilm | media-film | 3M |
| SimPlate | media | Idexx |
| Redigel | Media | RCR Scientific |
| ColiQuik | MUG/ONPG | Hach |
| LST-MUG | MPN media | Difco & GIBCO |
| CHROMagar | Medium | CHROMagar |
| E. coli | ||
| MUG disc | MUG | REMEL |
| CHROMagar | Medium | CHROMagar |
| EHEC | ||
| Rainbow Agar | Medium | Biolog |
| BCMO157:H7 | Medium | Biosynth |
| Fluorocult O157:H7 | Medium | Merck |
| Salmonella | ||
| Isogrid | HGMF | QA Labs |
| OSRT | Medium/ motility | Unipath (Oxoid) |
| Rambach | Medium | CHROMagar |
| MUCAP | C8esterase | Biolife |
| XLT-4 | Medium | Difco |
aAbbreviations: HGMF/MUG, hydrophobic grid membrane filter/4-methylumbelliferyl-β -D-glucuronide; ONPG, O-nitrophenyl β-D-galactoside; MPN, most probable number.
4. Conclusions
Bacterial pathogens and their toxins can cause illnesses such as diarrhea and spread through population. These pathogens are causing more and more outbreaks of disease every year. Therefore, rapid and reliable detection methods are needed. Conventional methods for the detection of enteric pathogen bacteria are sensitive. However, Traditional standard culture methods require long turnaround time for enrichment and confirmation of presumptive isolates and may require several days to obtain results. These methods are based on immunochemical and nucleic acid technologies and are alternatives for conventional methods, because these methods can provide results within hours. The DNA microarrays can facilitate whole genome comparisons among diverse strains and the identification of strain-specific and lineage-specific sequences.
The conventional PCR methods, automated fluorogenic, and quantitative real-time PCR kits have been invented and become available on the market. But in the laboratories that have lower sample throughput, commercialized automated immunoassay-based methods are less expensive. Optical techniques like SPR have better sensitivity, but they are expensive and complicated. The methods based on biosensor are rapid in detecting microbial pathogens within hours or even minutes. LAMP assays, peptide nucleic acid probes, DNA microarrays, and DNA chips are more advanced and potentially new rapid methods for enteric pathogens detection. Therefore, these rapid methods have become increasingly popular among laboratories and could be accepted as cost-effective and standard methods for pathogen detection in the future.