1. Background
Coagulase-negative staphylococci (CoNS) belong to normal microbial flora of the skin and mucous membranes. CoNS consists of a variety of staphylococci species and some of them such as Staphylococcus epidermidis and S. haemolyticus colonized permanently or transiently at the anterior nares, skin and mucous membranes, which may act as source of later bacteremia and other infections (1, 2). CoNS due to the expression of mecA gene specifies penicillin binding protein 2a (PBP2a), a transpeptidase with low affinity for β-lactams, which is typically resistant to methicillin (3, 4). The mecA gene is carried by staphylococcal cassette chromosome mec (SCCmec) (5). SCCmec is a group of mobile DNA elements that integrate into the chromosome of methicillin resistant S. aureus (6). In addition, the SCCmec contains the mec gene complex and the ccr gene complex. The mec gene complex consists of mecA, the regulatory genes and associated insertion sequences and has been classified into six different classes of A, B, C1, C2, D and E. Cassette chromosome recombinase (ccr) genes (ccrC or the pair of ccrA and ccrB) encode recombinases enzyme that act as a integration and excision of SCCmec into and from the chromosome (5, 7, 8).
The mechanism responsible for mecA transfer is not known, but data supports horizontal transfer of mec DNA between staphylococcal species and the mecA gene between different Gram-positive genera (7). According to the classes of the mec gene complex and the ccr gene types, 11 types (I to XI) of SCCmec have been shown for S. aureus (9). Methicillin resistance is common in CoNS and may play as an available reservoir of SCCmec for S. aureus to form methicillin resistant S. aureus (MRSA) (10). Due to the presence of new variants of ccr genes, SCCmec elements are different in MR-CoNS (9). In addition, high percent of SCCmec elements in MR-CoNS could not be typed using currently-available formats based on multiplex PCR (9-11). CoNS is usually much more resistant to antibiotics than S. aureus (12). There are no adequate data on the prevalence of methicillin resistance and SCCmec diversity among carriage strains of CoNS, particularly MR-CoNS in children. To find the information on SCCmec in nasal carriage MR-CoNS, 600 students were investigated.
2. Objectives
The aim of this study was to determine the prevalence of methicillin-resistant coagulase negative nasal colonization of staphylococci among healthy school students, to identify risk factors for colonization and characterize isolates using molecular and microbiological methods in Kashan, Iran.
3. Materials and Methods
3.1. Sample collection
The study was conducted on 7 to 19 years old students from February to May 2012 in Kashan, Iran. We collected samples from students with approval from the Ministry of education and data protection ability. The study was approved by the ethical committees of Kashan University of Medical Sciences (#2737). In addition, permission for sampling was asked from parents of students. A questionnaire was provided to each participant to collect demographic information, medical history and any factors that may be potentially related to CoNS nasal colonization and transmission. Nasal swab samples were obtained from both nares with sterile dry-cotton swabs, which directly inoculated onto blood agar and manitol salt agar.
3.2. Bacterial isolates
CoNS and Staphylococcusaureus were identified by conventional methods after incubation for 48 hours at 37ºC. Further evaluation by Gram staining, catalase, oxidase, clumping factor, tube Coagulase, heat stable nuclease, alkaline phosphatase, ornithine decarboxylase, novobiocin resistance, polymyxin B resistance, β-galactosidase, Voges-Proskauer, pyrrolidonyl aminopeptidase (PYR) and urease tests were performed on all colonies that their morphology was related to CoNS. Resistance to methicillin for all CoNS isolates was determined by Cefoxitin (30 μg) disk diffusion test (inhibition zone ≤ 24 mm for coagulase negative staphylococci except S. lugdunensis and inhibition zone ≤ 21 mm for S. lugdunensis) (13). Antimicrobial susceptibility tests: Antimicrobial susceptibility profiles were determined in accordance with the guidelines of the Clinical and Laboratory Standards Institute (CLSI) for MR-CoNS on Mueller-Hinton agar (6, 13). Used antibiotic disks were cefoxitin (30 μg), gentamicin (10 μg), erythromycin (15 μg), tetracycline (30 μg), clindamycin (2 μg), amikacin (30 μg), sulfamethoxazole (1.75 μg), trimethoprim (5 μg) and ciprofloxacin (5 μg) purchased from mast group Ltd., merseyside, UK.
Resistance to vancomycin was determined by E-test (liofilchem, Italy) in accordance with the guideline of the CLSI for MR-CoNS on Mueller Hinton agar (13). The S. aureus ATCC-29213 was used as the quality control at each set of tests. Carriage of MS-CoNS was not studied. DNA extraction: Frozen bacteria were sub-cultured onto 5% sheep blood agar plates prior to DNA extraction. For rapid DNA extraction, one to five bacterial colonies were suspended in 50 µL of sterile distilled water and heated at 99°C for 10 min. After centrifugation at 30000 g for 1 minute, 2 µL of the supernatant was used as template in a 25 µL PCR (14).
3.3. PCR amplification
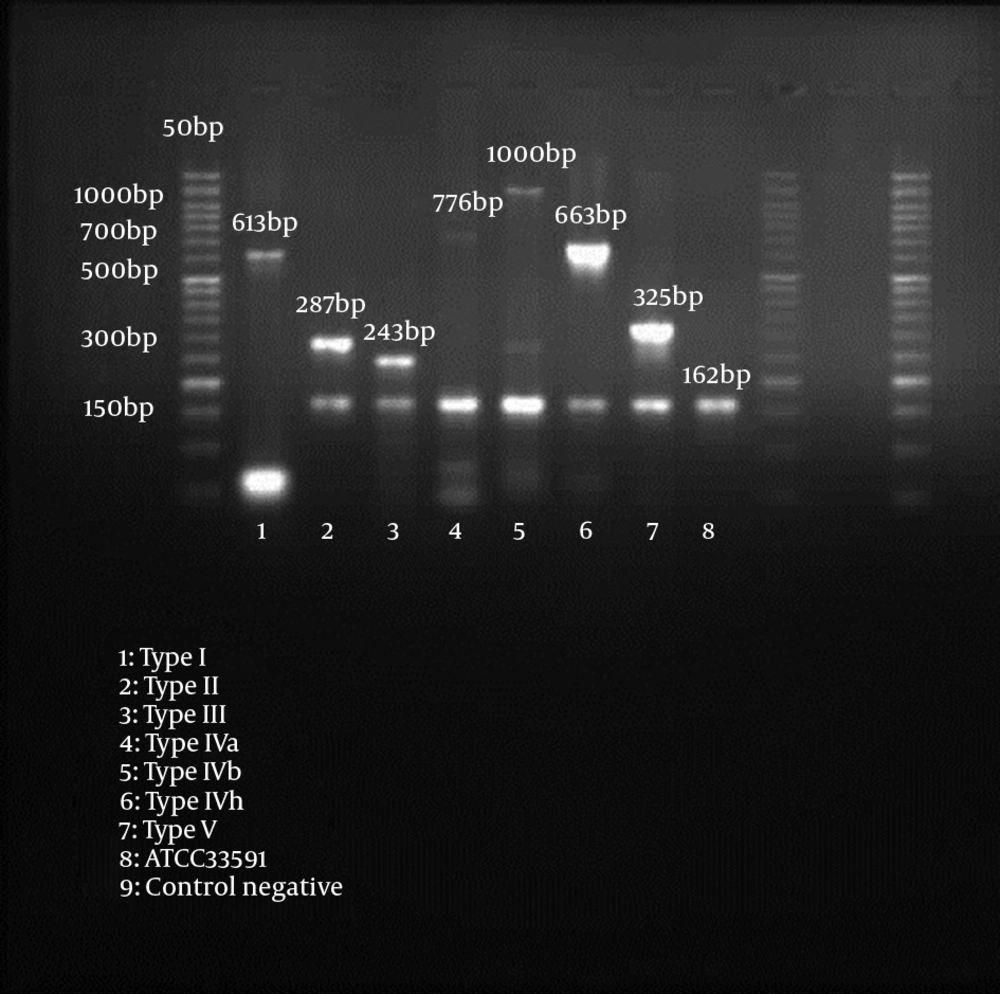
Methicillin resistance was detected by amplification of a 162 bp fragment from the mecA gene (15). Our SCCmec multiplex-PCR typing assay used for characterization of mec gene and ccr gene complexes, contained 4 primers each (mecI-F, mecI-R, IS1272-F and mecR1-R for mec gene M-PCR, and ccrAB-1, ccrAB-2, ccrAB-3, and ccrAB-4 for ccr gene M-PCR), respectively (14) (Table 1).
| Primer | Oligonucleotide Sequence (5’ - 3’) | Amplicon Size (bp) | Specificity | Reference |
|---|---|---|---|---|
| MecA-F | TCCAGATTACAACTTCACCAGG | 162 | mecA | (15) |
| MecA-F | CCACTTCATATCTTGTAACG | |||
| mecI-F | CCCTTTTTATACAATCTCGTT | 146 | Class A mec | (14) |
| mecI-R | ATATCATCTGCAGAATGGG | |||
| IS1272-F | TATTTTTGGGTTTCACTCGG | 1305 | Class B mec | (14) |
| mecR1-R | CTCCACGTTAATTCCATTAATACC | |||
| ccrAB-β2 | ATTGCCTTGATAATAGCCITCT | |||
| ccrAB-α2 | AACCTATATCATCAATCAGTACGT | 700 | Type 1 ccr | (14) |
| ccrAB-α3 | TAAAGGCATCAATGCACAAACACT | 1000 | Type 2 ccr | (14) |
| ccrAB-α4 | AGCTCAAAAGCAAGCAATAGAAT | 1600 | Type 3 ccr | (14) |
| ccrC-F | ATGAATTCAAAGAGCATGGC | 336 | Type 5 ccr | (14) |
| ccrC-R | GATTTAGAATTGTCGTGATTGC |
3.4. Statistical Analysis
Data was analyzed using SPSS (version 16.0 for Windows; SPSS Inc., Chicago, IL). Chi square or Fisher’s exact test was used for analysis of categorical data. A P value of < 0.05 was considered statistically significant.
4. Results
A total of 600 consecutive students including 261 male (43.5%) and 339 female (56.5%) were enrolled in this study. The overall prevalence of CoNS carriage was 71.7% (430 of 600). The mean age for the studied population was 12.73 years (± 3.466), which ranged from 7 to 19 years. Of 600 specimens being tested, 430 isolates (71.7%) were identified as coagulase-negative staphylococci (CoNS) and 109 (18.2%) as S. aureus. In addition, 42.2% of S. aureus were MRSA (46 of 109). CoNS consists various species as follows: S. lugdunensis 112 (26.1%), S. epidermidis 107 (24.9%), S. haemolyticus 105 (24.4%), S. saprophyticus 59 (13.7%), S. schleiferi 43 (10%) and S. simulans 4 (0.9%). A total of 72 (16.7%) MR-CoNS were identified. S. saprophyticus showed the highest methicillin resistance rate. Antibiotic susceptibility patterns of 72 carriage strains of methicillin-resistant coagulase-negative staphylococci (MRCoNS) are shown in Tables 2 and 3. The highest percent of MR-CoNS was susceptible to gentamicin (100%), amikacin (100%), ciprofloxacin (97.2%), clindamycin (93.1%), erythromycin (91.7%) and trimethoprim (88.8%), respectively. The lowest activity was found for sulfamethoxazole (83.3%). Approximately 7% of students had MRSA (30 of 430) and 23.6% of MRCoNS isolates had both MRSA and MRCoNS (17 of 72). The frequency rates for co-colonization presented as follows: MRSA + MR S. epidermidis 5.9% (1 of 17); MRSA + MR S. lugdunensis 23.5% (4 of 17); MRSA+ MR S. haemolyticus 35.3% (6 of 17); MRSA + MR S. saprophyticus 23.5% (4 of 17) and MRSA + MR S. schleiferi 11.8% (2 of 17). All of the MR-CoNS were susceptible to vancomycin (MIC: 0.17 to 0.75). Type IV was the most frequent SCCmec variant. Nineteen students (26.4%) were colonized by MRCoNS strains carrying SCCmec type IV, which five of them (26.3%) were identified as S. lugdunensis.
| Sccmec Types | Amikacin | Gentamicin | Ciprofloxacin | Vancomycin | Clindamycin | Erythromycin | Trimethoprim | Sulfamethoxazole |
|---|---|---|---|---|---|---|---|---|
| Type I No. 5 | 5 (100) a | 5 (100) | 5 (100) | 5 (100) | 4 (80) | 5 (100) | 3 (60) | 5 (100) |
| Type II No. 4 | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) |
| Type III No. 7 | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 6 (85.7) | 7 (100) |
| Type IV No. 19 | 19 (100) | 19 (100) | 18 (94.7) | 19 (100) | 19 (100) | 16 (84.2) | 18 (94.7) | 19 (100) |
| Type V No. 13 | 13 (100) | 13 (100) | 12 (92.3) | 13 (100) | 13 (100) | 12 (92.3) | 13 (100) | 13 (100) |
a Data is shown as No. (%).
| Species | Amikacin | Gentamicin | Ciprofloxacin | Vancomycin | Clindamycin | Erythromycin | Trimethoprim | Sulfamethoxazole |
|---|---|---|---|---|---|---|---|---|
| S. epidermidis (n = 9) | 9 (100) a | 9 (100) | 8 (88.9) | 9 (100) | 7 (77.8) | 9 (100) | 8 (88.9) | 8 (88.9) |
| S. lugdunensis (n =21) | 21 (100) | 21 (100) | 20 (95.2) | 21 (100) | 21 (100) | 18 (85.7) | 19 (90.5) | 18 (85.7) |
| S. haemolyticus (n = 17) | 17 (100) | 17 (100) | 17 (100) | 17 (100) | 16 (94.1) | 17 (100) | 15 (88.2) | 15 (88.2) |
| S. saprophyticus (n = 17) | 17 (100) | 17 (100) | 17 (100) | 17 (100) | 16 (94.1) | 15 (88.2) | 15 (88.2) | 13 (76.5) |
| S. schleiferi (n = 8) | 8 (100) | 8 (100) | 8 (100) | 8 (100) | 7 (87.5) | 7 (87.5) | 7 (87.5) | 6 (75) |
| Total | 72 (100) | 72 (100) | 70 (97.2) | 72 (100) | 67 (93.1) | 66 (91.7) | 64 (88.9) | 60 (83.3) |
a Data is shown as No. (%).
Among the 72 MR-CoNS, 48 isolates (66.7%) had single SCCmec type including type I (n = 5), II (n = 4), III (n = 7), IV (n = 19) and V (n = 13), whereas 5 isolates (6.9%) had two types including IV + V (n = 2), III + IV (n = 2), III + V (n = 1) (Table 4). In whole, 19 isolates (26.4%) were non-typeable SCCmec, because neither ccr genes nor mec genes could be amplified. SCCmec typing results for nasal carriage of methicillin-resistant coagulase-negative staphylococci are shown in Table 5.
| Staphylococcal isolate (s) | SCCmec type | ccr type | mec class | No. of Isolates |
|---|---|---|---|---|
| S. epidermidis (n = 9) | ||||
| I | Type 1 | Class B | 1 | |
| II | Type 2 | Class A | 0 | |
| III | Type 3 | Class A | 1 | |
| IV | Type 2 | Class B | 4 | |
| V | Type 5 | Class C | 2 | |
| Non-Typeable | 1 | |||
| S. lugdunensis (n = 21) | ||||
| I | Type 1 | Class B | 1 | |
| II | Type 2 | Class A | 1 | |
| III | Type 3 | Class A | 2 | |
| IV | Type 2 | Class B | 5 | |
| V | Type 5 | Class C | 3 | |
| IV + V | Type 2 + 5 | Class B + C | 1 | |
| III + IV | Type 3 + 2 | Class A + B | 1 | |
| Non-Typeable | 7 | |||
| S. haemolyticus (n = 17) | ||||
| I | Type 1 | Class B | 1 | |
| II | Type 2 | Class A | 2 | |
| III | Type 3 | Class A | 1 | |
| IV | Type 2 | Class B | 3 | |
| V | Type 5 | Class C | 4 | |
| III +V | Type 3 + 5 | Class A + C | 1 | |
| IV + V | Type 2 + 5 | Class B + C | 1 | |
| Non-Typeable | 4 | |||
| S. saprophyticus (n = 17) | ||||
| I | Type 1 | Class B | 1 | |
| II | Type 2 | Class A | 1 | |
| III | Type 3 | Class A | 2 | |
| IV | Type 2 | Class B | 4 | |
| V | Type 5 | Class C | 3 | |
| III + IV | Type 3 + 2 | Class A + B | 1 | |
| Non-Typeable | 5 | |||
| S. schleiferi (n = 8) | ||||
| I | Type 1 | Class B | 1 | |
| II | Type 2 | Class A | 0 | |
| III | Type 3 | Class A | 1 | |
| IV | Type 2 | Class B | 3 | |
| V | Type 5 | Class C | 1 | |
| Non-Typeable | 2 |
| MR-CoNS Characteristic | Positive | Negative | Total | P Value |
|---|---|---|---|---|
| Age, y | 0.284 | |||
| 7 - 11 | 20 (27.8) a | 134 (37.4) | 154 (35.8) | |
| 12 - 15 | 28 (38.9) | 123 (34.4) | 151 (35.1) | |
| 16 - 19 | 24 (33.3) | 101 (28.2) | 125 (29.1) | |
| Gender | 0.452 | |||
| Male | 30 (16.2) | 155 (85.8) | 185 (43) | |
| Female | 42 (17.1) | 203 (82.9) | 245 (57) | |
| Previous antibiotic usage | 0.088 | |||
| Yes | 7 (9.7) | 60 (16.8) | 67 (15.6) | |
| No | 65 (90.3) | 298 (83.2) | 363 (84.4) | |
| Species | 0.021 | |||
| S. epidermidis | 9 (12.5) | 98 (27.4) | 107 (24.8) | |
| S.lugdunensis | 21 (29.2) | 91 (25.4) | 112 (26.1) | |
| S. haemolyticus | 17 (23.6) | 88 (24.6) | 105 (24.4) | |
| S. saprophyticus | 17 (23.6) | 42 (11.7) | 59 (13.7) | |
| S. schleiferi | 8 (11.1) | 35 (9.8) | 43 (10) | |
| S. simulans | 0 (0) | 4 (1.1) | 4 (1) |
a Data is shown as No. (%).
5. Discussion
The prevalence of CoNS nasal carriage among children participating in this study was as high as 71.7%. Few surveys performed to examine CoNS nasal colonization in children. We found that the prevalence of MR-CoNS was 16.7%. There are a very limited number of reports on MR-CoNS nasal carriage among children. In total, SCCmec types were determined for most isolates (53 of 72), the remaining 26.4%, ccr and mec complexes could not be amplified in any way. In agreement with previous reports, types I and II SCCmec were few, while type IV was relatively common (4, 6). In this study, type IV was the most common type presented in 19 of 72 MR-CoNS isolates either alone or combined with other types, followed by type V (13 of 72) and then III (7 of 72).
The circulation of different types of SCCmec in MR-CoNS varies according to the host species and geographical locations. In this study, SCCmec type IV mainly presented in S. lugdunensis and S. epidermidis and type V dominated in S. haemolyticus. Recent data from Japan confirmed that SCCmec IVa has been found in most community-acquired methicillin-resistant S. epidermidis (16). Other researchers reported that 36% of methicillin-resistant Staphylococcus epidermidis carries a SCCmec type IV-like structure (17). SCCmec types III, IV and V were common in MR-CoNS and several isolates can harbor more than one type. Epidemiological studies and molecular characterization of methicillin-resistant staphylococci from healthy Jordanian population showed the MR-CoNS carriage at 54.2% and the most common isolates were S. epidermidis SCCmec type Iva (18). Another report indicated that SCCmec V was predominant in methicillin-resistant S. haemolyticus and the mec complex class C was the most common (19).
Co-existence of two SCCmec elements in MR-CoNS was widespread (9). Other studies showed that SCCmec types with the traditional PCR SCCmec typing method (including mec and ccr gene complex typing) in a considerable proportion of MR-CoNS isolates could not be assigned by currently-available PCR-based methods (9, 18, 20). In a report from Miami, Florida, presence of non-typeable elements signified large challenges for SCCmec typing in MR-CoNS (21). In accordance with the available data, SCCmec elements are more diverse in MR-CoNS, by new variants of ccr genes (8). Moreover, many SCCmec elements in MR-CoNS could not be typed using currently-available schemes based on multiplex PCR (22). Additional investigations, including sequencing the mec element, are needed to characterize these currently not-typeable isolates.
Previous studies also have shown variations in SCCmec cassettes. These variations include strains containing both SCCmec type IV and ccrC, strains carrying multiple ccr genes, strains carrying ccr genes without a mec complex and a mecA-positive MRSA strain with neither ccr genes nor a mec complex (23). In this study, co-colonization of MRSA and MRCoNS showed in 23.6% of isolates. Several researches reported that SCCmec transfer from MR-CoNS to methicillin-susceptible S. aureus could occur, although its mechanism remains unknown (7, 10). More than 90% of MR-CoNS were susceptible to gentamicin, amikacin, clindamycin, ciprofloxacin and erythromycin. All of MR-CoNS were sensitive to vancomycin. Indeed, coagulase-negative staphylococci were the first organisms in which acquired resistance to glycopeptides antibiotics was reported, but vancomycin resistance in Coagulase-negative staphylococci is still uncommon.
All 21 S. lugdunensis isolates analyzed in our study had favorable antibiotic susceptibility patterns; they were completely sensitive to amikacin, gentamicin, vancomycin and clindamycin. Interestingly, all S. haemolyticus isolates were sensitive to amikacin, gentamicin, ciprofloxacin, vancomycin and erythromycin. In S. epidermidis, phenotypic clindamycin resistance was significantly more common than other MR-CoNS species. Approximately, 40% of type I MR-CoNS was resistance to trimethoprim. From the studied risk factors, there was no significant association between sex, age and previous antibiotic usage. Distribution of MRCoNS in persons with no causal risk factor has been reported in other studies (12, 16). Inappropriate and non-sophisticated antibiotic therapy may inhibit normal sensitive bacterial flora of the body and provides an environment, which facilitates colonization by antibiotic-resistant bacteria. Although an individual who receives antibiotic does not become colonized by methicillin-resistant bacteria except he or she gets in contact with these bacteria. The emerging spread of community-acquired MR-CoNS strains threatens public health, suggesting that students are potentially the most important reservoirs of methicillin resistance in community. This study provided the first description of MR-CoNS in healthy students in Iran. In conclusion, we found high rates of MRSA and MR-CoNS carriage among students in an area with a high rate of antibiotics use.
This study had some limitations. The method used in this study for SCCmec classification of CoNS was derived from the S. aureus prototype. Due to this limitation, a large number of cases could not be typed. According to our results, close monitoring of MR-CoNS epidemiology in community is required to estimate contribution of hidden MRSA reservoirs. A systematic understanding of the molecular epidemiology of MR-CoNS is necessary for efficient detection, treatment, control and prevention of diseases caused by this organism.