1. Background
Acinetobacter calcoaceticus baumannii (ACB) complex are coccobacillus Gram-negative bacteria. They are opportunistic and associated with some health-related infections (1, 2). Such bacterial infections, particularly in patients hospitalized at intensive care units, are very dangerous (3). In addition, colonization rates are rising among hospitalized patients, especially those hospitalized for a long time or those who are on extended antibiotic therapy or receiving anti-cancer treatment (2, 4). This organism can infect the respiratory tract, blood, soft tissues, urinary tract and the central nervous system. Acinetobacter calcoaceticus baumannii complex has emerged as a highly troublesome pathogen in hospitals, and is resistant to multiple antimicrobial agents (5).
The various species of Acinetobacter are mostly intrinsic with acquired resistance to common antimicrobial agents, thus, the treatment of respective infections is very difficult (6). In the recent years, there have been reports on the incidence of Acinetobacter infections with Multi-Drug Resistance (MDR) around the world (7). Excessive use of antimicrobial drugs in the treatment of such infections has caused problems in treatments leading to growing concerns in many communities. Health communities are concerned about the resistance of ACB complex to all conventional antibiotics, even to broad-spectrum antibiotics such as carbapenems and sulbactam (8). At present, these organisms only have little resistance to colistin and polymyxin B.
Keeping the spread of Acinetobacter under control is very difficult because these organisms disseminate easily in hospital environments (8). Epidemiological studies of nosocomial infections are essential to prevent these infections. However, phenotypic methods such as antibiotic resistance tests, because of phenotypic characterization variation and low differentiation power, are not sufficient alone in epidemiological studies (9). To obtain more reliable results, genotypic methods should be used along with phenotypic characterization. Pulsed Field Gel Electrophoresis (PFGE) is the most widely used method for valuable typing of epidemiological studies in different regions and many researchers have used this method to evaluate the similarities between the strains of ACB complex (8, 10-13). Comparison of pulse-field patterns is a useful method for obtaining strain relationships in a certain species.
2. Objectives
Pulsed Field Gel Electrophoresis was used in this study as a reliable method for differentiating between the strains of ACB and distinguishing respective clonal relationships, as well as the relationship between genetic and phenotypic patterns. In addition, the present study was designed for the isolation of strains from clinical samples, identification using appropriate methods and evaluation of their antibiotic resistance patterns. In doing so, a logical approach could be proposed to treat infected patients.
3. Patients and Methods
3.1. Patients and Bacterial Isolation
During a period of nine months in 2011, 117 isolates were collected from the samples of patients with suspected Acinetobacter infections, who were referred to Nemazee hospital of Shiraz, southern Iran. All presumptive Acb complexes, which were Gram-negative coccobacilli, oxidase negative and non-lactose fermentative, were identified at the species level by using biochemical test and conventional analytical profile index (API) (Microgen GNA-IDTM) (14). As the cases enrolled in this study were only patients with nosocomial infections, the inclusion criteria were antibiotic use during the last 15 days and infections noted 48 hours after admission or within four weeks after a previous discharge (15). Informed written consents were obtained from all patients. The design and protocol of the study were also approved by the ethics committee of Professor Alborzi Clinical Microbiology Research Center.
3.2. Antibiotic Susceptibility Patterns
Susceptibility of all isolates to different antibiotics was determined by the disk diffusion method, as recommended by the National Committee for Clinical Laboratory Standards (CLSI), (16) with commercial antimicrobial disks (Mast. Co. UK). The antibiotics consisted of: gentamicin (GM), ticarcillin (TC), ceftazidime (CAZ), co-trimoxazole (TS), imipenem (IMI), piperacillin tazobactam (PTZ), amikacin (AK), aztreonam (ATM), sulbactam (SAM), meropenem (MEM), tobramycin (TN), cefotaxime (CTX), ceftriaxone (CRO), colistin (CO) and polymyxin B (PB). Escherichia coli ATCC 25922 was used for quality control purposes in antibacterial susceptibility determination.
3.3. Pulsed-Field Gel Electrophoresis
In this study, we used the Seifert procedure with some modifications (17). The isolates were purely cultured on Mueller-Hinton medium. Subsequently, the colonies were mixed with 1 cc of buffer suspension (10 mM tris HCl [pH 8], 100 Mmol ethylenediaminetetraacetic acid (EDTA)). Next, 1 cc of 2% Low Melting Point Agarose (LMA) was added to the tube containing the microbial solution. The resulting mixture was transferred to a gel mold until the gel was formed completely. Then the gel was transferred to the flask to which we added 25 cc of lysing buffer (50 mM tris HCl [pH 8], 50 Mmol EDTA, 1% lauroylsarcosine), and 125 µ of 20 mg/cc proteinase K (Fermentase, Lithuania)) so that the gel floated on the solution. The resulting solution was shaken by a shaker at 54°C for 18 hours. Next, each DNA plug was initially washed with sterile distilled water, and then, with sterile Tris EDTA washing buffer (10 mM tris HCl [pH8], 1 mmol/liter EDTA) on an incubated shaker at 50°C. Subsequently, the DNA plug was transferred to a sterile Eppendorf tube and preserved in the heater for 18 hours at 37°C (Takara Biotechnology, Dalian, CO, LTD). DNA preparations were transferred to the wells of a nucleic acids agarose (molecular grade, Amersham Bioscience, Sweden). Next, the buffer tank containing EDTA, boric acid and Tris-base at pH 8 was prepared for isolating bacterial genome and transferred to the PFGE instrument (Gene Navigator System, Amersham Bioscience, Sweden). Electrophoretic conditions used, were as follows: initial switch time of five seconds; second switch time of 20 seconds; final switch time of 40 seconds; temperature of 6°C, run time of 33 hours; angle of 120° and gradient of 6 v/cm. In each set a 1000 bp-lambda ladder (Biolabs, New England) was used as a molecular marker.
PhotoCapt software (Vilber Loumart, Marnesla Valle, France) was used to determine the molecular weights of the sample profiles. The bands were evaluated according to the DNA marker and were scored across all samples. They were recorded as number one for present or zero for absent. Subsequently, the data were used to calculate pair-wise similarity coefficient following Jaccard’s method. To generate a dendrogram using the average linkage procedure, the analysis of the similarity coefficients matrices was performed using Unweighted Pair-Group Method Analysis (UPGMA). To calculate correlations among the variables, standardized data matrices were used. The correlations were subjected to Eigenvector analysis to evince the first three uttermost elucidative principal components. To study the patterns of variations observed among the isolates, the three principal components were plotted. The Numerical Taxonomy System (NTSYS) pc software version 2.02i (Exeter software, New York) was used to conduct all the numerical analyses.
4. Results
4.1. Patients and Bacterial Isolation
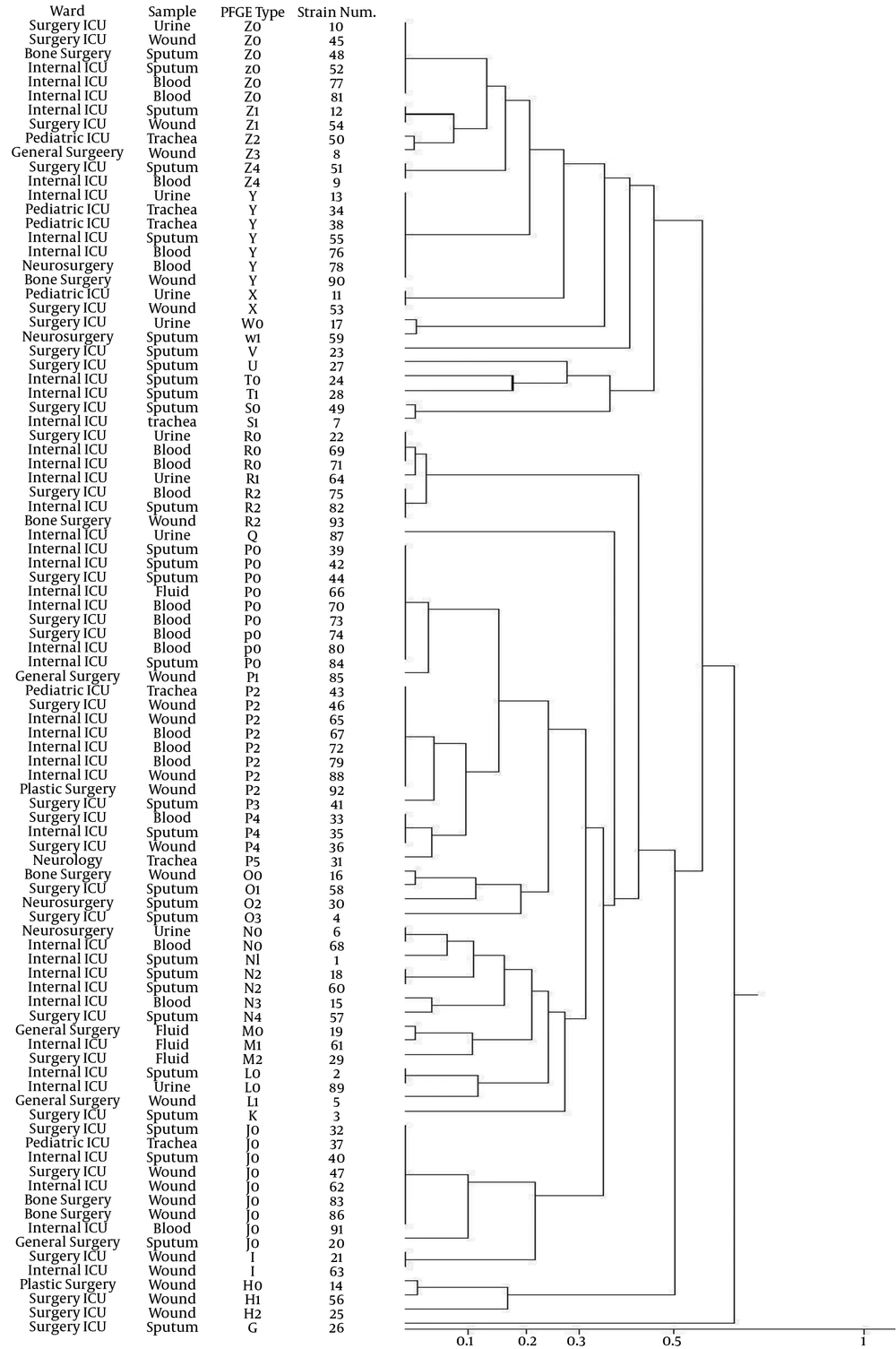
In total, 117 samples from patients aged between 16 days and 95 years, with suspected ACB complex infection were obtained; of which 93 samples were ACB complex while others were Gram-negative organisms (such Pseudomonas and E. coli). Among ACB complex isolates, 31 (33.4%) samples were from sputum and throat secretions, 23 (24.8%) from wound, 19 (20.4%) from blood cultures, nine (9.6%) from urine, seven from trachea (7.6%), and four from body fluids (4.2%). Samples were collected from different wards of Nemazee Hospital with corresponding distributions presented in Figure 1. To avoid multiple entries from a single patient, only the first positive Acinetobacter culture for a given patient was included.
4.2. Antibiotic Susceptibility Patterns
Antibiotic sensitivity patterns of the isolates were determined by the standard disc diffusion method and the results are shown in Table 1. High resistance was observed among the tested antibiotics, except CO and PB. Over 90% of the strains exhibited resistance to antibiotics CTX, TS, CRO, ATM, CAZ and TC.
| Antibiotics | Total Resistant No. (%) |
|---|---|
| Cefotaxime (CTX) | 92 (98.9) |
| Co-trimoxazole (TS) | 90 (96.7) |
| Ceftriaxone (CRO) | 89 (95.7) |
| Aztreonam (ATM) | 87 (93.5) |
| Ceftazidime (CAZ) | 86 (92.5) |
| Ticarcillin (TC) | 85 (91.4) |
| Piperacillin Tazobactam (PTZ) | 81 (87.1) |
| Amikacin (AK) | 77 (82.8) |
| Meropenem (MEM) | 71 (76.3) |
| Gentamicin (GM) | 67 (72) |
| Imipenem (IMI) | 65 (69.9) |
| Tobramycin (TN) | 64 (68.8) |
| Sulbactam (SAM) | 54 (58.1) |
| Polymyxin B (PB) | 2 (2.2) |
| Colistin(CO) | 1 (1.1) |
Antibiotic Resistance of the Ninety-Three Acinetobacter calcoaceticus baumannii Complex Strains, Isolated From Patients With Nosocomial Infections, in Different Wards of Nemazee Hospital a
Ninety-eight percent of the isolates were resistant to three or more antibiotics and were designated as MDR (18). Thirty-five patterns of resistance were recognized for the ACB complex strains (Table 2) and the most common pattern was resistance to 13 antibiotics; ATM, AK, CRO, PTZ, TC, SXT, IMI, GM, CAZ, MEM, TOB, CTX and SAM. This pattern was repeated among 21 strains, followed by pattern No. 7, which was seen amongst 18 strains. Both patterns were the dominant patterns in the Emergency Coronary Unit (ICU) Internal patients.
| No. | Antibiotic Resistant Patterns | Frequency |
|---|---|---|
| 1 | ATM, AK, CRO, PTZ, TC, CO, SXT, PB, IMI, GM, CAZ, MEM, TOB, CTX, SAM | 1 |
| 2 | ATM, AK, CRO, PTZ, TC, SXT, IMI, GM, CAZ, MEM, TOB, CTX, SAM | 21 |
| 3 | ATM, AK, CRO, TC, SXT, PB, IMI, GM, CAZ, MEM, TOB, CTX, SAM | 1 |
| 4 | ATM, AK, CRO, PTZ, TC, SXT, GM, CAZ, MEM, TOB, CTX, SAM | 4 |
| 5 | ATM, AK, CRO, PTZ, TC, SXT, IMI, GM, CAZ, MEM, CTX, SAM | 8 |
| 6 | ATM, AK, CRO, PTZ, TC, SXT, IMI, CAZ, MEM, TOB, CTX, SAM | 1 |
| 7 | ATM, AK, CRO, PTZ, TC, SXT, IMI, GM, CAZ, MEM, TOB, CTX | 18 |
| 8 | AK, CRO, PTZ, TC, SXT, IMI, GM, CAZ, MEM, TOB, CTX | 1 |
| 9 | AK, CRO, TC, SXT, IMI, GM, CAZ, MEM, TOB, CTX, SAM | 1 |
| 10 | CRO, PTZ, TC, SXT, IMI, GM, CAZ, MEM, TOB, CTX, SAM | 1 |
| 11 | ATM, AK, CRO, PTZ, TC, SXT, GM, CAZ, TOB, CTX, SAM | 3 |
| 12 | ATM, AK, CRO, PTZ, TC, SXT, GM, CAZ, MEM, CTX, SAM | 1 |
| 13 | ATM, AK, CRO, PTZ, TC, SXT, IMI, CAZ, MEM, CTX, SAM | 5 |
| 14 | ATM, AK, CRO, PTZ, TC, SXT, IMI, GM, CAZ, MEM, CTX | 3 |
| 15 | ATM, AK, CRO, PTZ, TC, SXT, IMI, CAZ, MEM, TOB, CTX | 1 |
| 16 | CRO, PTZ, SXT, IMI, GM, CAZ, MEM, TOB, CTX, SAM | 1 |
| 17 | ATM, AK, CRO, PTZ, TC, SXT, CAZ, MEM, TOB, CTX | 1 |
| 18 | ATM, AK, CRO, PTZ, TC, SXT, IMI, CAZ, MEM, CTX | 1 |
| 19 | ATM, AK, CRO, PTZ, TC, SXT, CAZ, TOB, CTX | 1 |
| 20 | ATM, CRO, PTZ, TC, SXT, GM, CAZ, TOB, CTX | 1 |
| 21 | CRO, TC, SXT, IMI, GM, CAZ, MEM, TOB, CTX | 1 |
| 22 | ATM, AK, CRO, TC, SXT, GM, TOB, CTX, SAM | 1 |
| 23 | ATM, CRO, PTZ, TC, SXT, CAZ, TOB, CTX, SAM | 1 |
| 24 | ATM, CRO, PTZ, TC, SXT, CAZ, TOB, CTX | 1 |
| 25 | ATM, AK, CRO, SXT, CAZ, TOB, CTX, SAM | 1 |
| 26 | ATM, AK, CRO, PTZ, TC, SXT, CAZ, CTX | 1 |
| 27 | ATM, AK, CRO, PTZ, TC, SXT, CTX | 1 |
| 28 | ATM, CRO, PTZ, TC,CAZ, CTX, SAM | 3 |
| 29 | ATM, CRO, SXT, CAZ, TOB, CTX | 1 |
| 30 | ATM, TC, SXT,CAZ, TOB, CTX | 1 |
| 31 | ATM,CRO, PTZ, TC, SXT, CTX | 1 |
| 32 | ATM, AK, CRO, SXT, CAZ, CTX | 1 |
| 33 | ATM, CRO, SXT, CTX | 1 |
| 34 | ATM, SXT, CTX | 2 |
| 35 | SXT | 1 |
| 36 | Total | 93 |
Antibiotic Resistance Patterns of Acinetobacter calcoaceticus baumannii Complex Clinical Isolates, Isolated From Patients With Nosocomial Infections, in Different Wards of Nemazee Hospital a
4.3. Pulsed-Filed Gel Electrophoresis
Using this technique and considering the designed dendrogram (Figure 1), in total, 47 genetic patterns were obtained and assigned letters from Z to G. With cut off level 80%, clone P (P1, P2, P3, P4, P5) holds the largest number of organisms. Pattern P1 with nine identical isolates was the most repeated pattern. Eleven samples with strains of clone P were isolated from patients in the internal ICU, eight from ICU surgical, and one sample from general ICU, pediatric, plastic surgery and neurology each. Band sizes obtained by PFGE ranged from 6 to 590 Kbp and band number ranged from 8 to 18. The most frequent patterns were those with 12 and 13 bands detected in 24.7% and 20.4% of the strains, respectively. The patterns with 8 and 18 bands accounted for 1.1% of the total isolated strains.
5. Discussion
Acinetobacter has been one of the main causes of nosocomial infections in the recent years. These opportunistic bacteria are the second agent isolated from hospitalized patients after Pseudomonas strains. In a report published in 2004 by the Center for Disease Control and Prevention (CDC), A. baumannii was the cause of 80% of infections (19). In the present study, based on biochemical tests and using standardized methods, 100% of the isolates were found to be ACB complex, which is almost consistent with the results of other studies (17, 20). Acinetobacter baumannii is the most common isolate obtained from the skin, blood, sputum, pleural fluid and urine (21). As many studies have shown, the respiratory tract is the body site where the organism is most frequently isolated in the ICU (22-25). In this study, the majority of studied patients were hospitalized in the ICU and most samples were collected from throat and sputum cultures and the rest of the samples were isolated from wound, blood, urine and tracheal fluid.
Nosocomial infections caused by the strains of these bacteria are increasingly becoming resistant to a range of antibiotics. For example, previous Iranian studies on resistance to MEM and IMI reported lower resistance rates than that of our study (26, 27). In another study conducted in Turkey (26), our neighboring country, all the samples were found to be sensitive to these antibiotics. As seen in Table 1, among the ACB complex except against CO and PB, high rates of antibiotic resistance were observed, indicating that these two antibiotics could be the most effective antibacterial agents against the respective infections. Similarly, there have been several reports of resistance to these antibiotics (28, 29), with some being alarming (30). In some other studies, resistance to CO was lower than that in our study (31, 32). According to the present study, CO was the least administered antibiotic, i.e., out of the 93 patients, only seven patients received this antibiotic, and consequently the least resistance to this antimicrobial agent was observed.
Although in the present study, resistance to PTZ was high, the highest resistance was observed to third generation cephalosporins, including TS, ATM, and TC (90%). In some studies, high percentage of resistance to TS, PTZ, CTX and CAZ was observed among the examined isolates (33, 34). Resistance to TN and SAM in the present study was comparable to the rates reported by previous studies from Iran (26, 27). In this study, a total of 35 phenotypic patterns were observed among the 93 strains, of which the predominant pattern was repeated in 21 strains, which was resistance to 13 antibiotics. Different patterns have been demonstrated in previous studies (33-36). Appearance of varying resistance patterns in different nations might be due to factors including financing policies, doctors and patients visiting and prescribing habits (37).
Increase of MDR organisms has led to concern in the medical community. There are many causes responsible for emerging MDR ACB complex infections in hospitals including length of hospitalization and staying at the ICU, which was an average of 24 days in the present study. Given the easy spread of Acinetobacter and its ability to survive in the environment for 13 days (19), there was enough time for the transmission of infection. Other factors such as number of hospital beds, and invasive and non-invasive procedures performed on the patients at the ICU, could be the cause of emerging MDR ABC strains. Since the majority of patients in this study were from the ICU with invasive procedures, the presence of MDR ABC was not unexpected. The majority of samples were isolated from the respiratory tract, therefore, it could be suggested that the methods associated with mechanical ventilation can facilitate the transmission of MDR ABC amongst patients in a ward.
In this study, similar to previous reports (30, 31, 38), a high prevalence of MDR isolates was found among the samples (98.9%). One of the reasons for the selection of ABC in hospital epidemics is the outbreak of MDR caused by this organism, as clearly demonstrated by previous studies (5, 39). The results of PFGE and similarity of genetic patterns of MDR strains confirmed this point. A specific phenotypic pattern was not observed in the epidemic clone P. As observed, most strains in this clone exhibited resistance to all aminoglycosides and carbapenems. Simultaneous presence of resistance to the two types of antibiotics can give rise to increasing epidemic infection in the studied hospital. Since these two antibiotics were the most widely used agents to treat suspected patients in the studied hospital, it can be suggested that based on the selective pressure related to antibiotic exposure, clonally related strains of Acinetobacter with different susceptibility patterns can be simultaneously traced in a hospital.
In many countries, various studies have shown genetic similarity among most strains of A. baumannii in a single hospital or different hospitals (13, 31, 40). Indeed, isolates with the same clonality have caused epidemics. In the present study, using the PFGE method as the gold standard typing method for Acinetobacter (31), we obtained 47 patterns of which type P with 23 strains by cut off level of 80%, included most strains with the same genetic pattern. Using this method, numbers of DNA molecular bands were between 8 and 18 and bands ranged in size from 6 to 590 Kbp. Changes in the genetic diversity of strains in different geographical areas can cause different patterns of bands in different laboratories.
A close genetic relationship among ABC strains showed the distribution of organisms in the studied hospital environment. Regarding clone P, most isolates were separated from ICUs and three phenotypic profiles of No.2, 5 and 7 were observed among the strains. In other words, similar a genetic pattern does not necessarily imply a similar phenotypic pattern. It can be stated that the mechanisms of plasmids resistance and changes in gene regulation or point mutations are indistinguishable by PFGE. In some clones, observations were different, i.e. strains with different genetic patterns had similar phenotypic patterns, which can be attributed to plasmid or transposons that can carry multiple resistance genes and then make the strains resistant to multiple antibacterial agents simultaneously, and due to the different genetic mechanisms, different strains have similar phenotypic patterns.
With respect to the above mentioned findings, identifying the bacterial resistance patterns of ABC in various hospitals over a specified time period seems to be necessary. As revealed by the present study, all the investigated Acinetobacter isolates were highly resistant to the examined antibiotics because of unwise and untimely use of antibiotics to treat infections. In view of the easy spread of ABC in nosocomial infections, accurate identification, antibiotic patterns evaluation and updating infection control procedures can contribute to the selection of more appropriate antibiotic therapy programs. The results of PFGE and the presence of closely related bacterial strains suggest that preventive measures such as education of clinicians, patients and visitors, hand hygiene, isolation of patients, periodic culture of patients and staff, changes in antibacterial usage, and regular disinfection of medical devices particularly for invasive procedures are warranted.