1. Background
Hepatitis C virus (HCV) is a single stranded RNA virus belonging to Flaviviridae family. HCV is a global health problem with an estimated of 3% infection of the world’s population. It was first discovered in 1989 and initially known as non-A, non-B hepatitis (NANBH). This virus primarily affects the liver and causes several severe diseases such as chronic hepatitis, cirrhosis, and hepatocellular carcinoma in human. The virus may transmit via intravenous drug usage, blood transfusion, organ transplantations, sexual contacts, and vertical transmission. The combination of pegylated IFN-α and ribavirin, with either boceprevir or telaprevir in some cases are administered to patients (1-5). Despite many studies related to this virus, no effective vaccine has been developed yet. This is partly due to the lack of suitable laboratory animal models and cell culture system that supports the viral replication. Hepatocytes and lymphocytes are permissive to HCV infection in vivo. However, in vitro condition, the virus is capable of primarily infecting these cells but cannot complete its propagation (6-8). Normally, this virus cannot replicate in cell culture systems. Only some limited HCV isolates such as H77 (genotype 1a), CG1b (genotype 1b), and JFH1 J6 (genotype 2a) are capable of replicating in Huh-7 cells.
JFH1 is a strain of HCV that was originally isolated from a Japanese patient with fulminant hepatitis. Transfection of the JFH1 genomic RNA into Huh-7 cell line leads to the production of HCV particles that are infectious in chimpanzee and can also replicate in vitro without adaptive mutations, thus producing infectious particles (8-11). Indeed, the establishment of an effective cell culture system for various genotypes and strains of HCV provides a valuable tool for studying the pathogenesis of HCV, and development of new antiviral agents and vaccines against this virus. In the present study, JFH1 isolate of HCV was cultured in Huh-7.5 cells. Subsequently, viral load of infectious viruses was evaluated by real-time PCR.
2. Objectives
The present study aimed to evaluate the multiplication of hepatitis C virus in a cell culture system and determination of its viral load.
3. Materials and Methods
3.1. Cells
Huh-7.5 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen, Canada), 0.2% sodium bicarbonate (Sigma, USA), 100 U/mL penicillin (Gibco, UK), and 100 µg/mL streptomycin (Gibco, UK) at 37°C in a 5% CO2 atmosphere.
3.2. Plasmid Preparation
JFH1 vector was transferred into Escherichia coli strain JM109 and the plasmid was purified from an overnight culture of recombinant E. coli by using QIAprep Miniprep Kit (Qiagen, USA). The quality and concentration of purified plasmid DNA were controlled using agarose gel electrophoresis together with measuring Optical Density (OD) of DNA at wave lengths of 260 nm and 280 nm.
3.3. DNA Digestion and RNA Synthesis
Double stranded linear DNA with blunt ends was used as a template for in vitro transcription. About 40 µg of JFH1plasmid was digested by Xba1 restriction enzyme (Fermentas, Germany). Mung bean nuclease was used to obtain blunt ends in digested plasmids. To eliminate the enzyme, phenol/chloroform protocol was applied. Briefly, 400 µL of a 1:1 phenol/chloroform mixture was added to the sample. After mixture, it was centrifuged at14000 rpm for 5 minutes. Aqueous phase was transferred to a new tube. Equal volume of chloroform was added, mixed and centrifuged at14000 rpm for 5 minutes. Then, aqueous phase was transferred into a new tube and equal volume of isopropanol was added. The tube was incubated for 30 minutes at 20°C. After incubation, it was centrifuged at14000 rpm for 45 minute at 4°C. Pellet was washed in 300 µL of 70% cold ethanol by centrifugation at 14000rpm for 10 minutes. Ethanol was discarded and pellet was air dried. The final pellet was dissolved in 20 μL of RNase-free water.
For RNA synthesis, transcription mixture consisted of 5 μL of Transcription buffer (5X), 5 μL of 10 mM ATP/GTP/CTP/UTP Mixture, 1 μg of linearized template DNA, 0.5 μL of (20U) RNase inhibitor, 1.5 μL of (30U) T7 Polymerase, and RNase-free water up to 25 μL. The mixture underwent overnight incubation at 37°C. Following incubation, to eliminate the template DNA, 2U of DNase I was added to the mixture and was incubated at 37°C for 15 minutes. To inactivate DNase enzyme, 2 μL of 0.5 M EDTA (pH 8.0) was added and the mixture was incubated at 65°C for 10 minutes. The final concentration of synthesized RNA was determined by gel electrophoresis and OD measurement.
3.4. RNA Transfection
One day before transfection, 3 × 105 Huh-7.5 cells in 2 mL of DMEM media were cultured in a 6-well plate and incubated at 37°C under 5% CO2 condition. After reaching 80% confluency, the growth medium was removed from cells and washed for one time with 2 mL of Opti-media (Invitrogen, Canada). In the next step, it was incubated at 37°C under 5% CO2 condition for 30 minute. The transfection mixture was prepared in DMRIE-C (Invitrogen, Canada) according to the manufacturer’s instructions. About 48 hours after transfection, culture media was collected for the following steps.
3.5. Infection of the Cells
One day before infection, 1 × 104 Huh-7.5 cells were cultured in a 6-well plate as described in the previous section. One milliliter of supernatant from transfected cells was used for the infection of the cells. After 1.5 hours incubation, the cells were kept at 37°C, under 5% CO2 condition for the virus to be adsorbed. Supernatant was discarded and cells were washed 3 times with phosphate buffer saline (PBS). Complete DMEM, containing 10% FBS was added and the cells were incubated for 48 h at 37°C under 5% CO2 condition. After incubation period, cells were washed 3 times for the second time with PBS. Next, fresh DMEM, containing 10% FBS was added and cells were incubated for more than 48 hours. Supernatant of infected cells was collected and analyzed by real-time PCR.
3.6. Viral Load Analysis by Real-Time PCR
The RNA copy number of hepatitis C virus (HCV) in supernatant of infected cells was determined by using HCV quantitative Real Time PCR Kit (Professional Biotech Pvt. Ltd, India). Briefly, viral RNA was isolated from culture media using QIAamp Viral RNA Mini Kit (Qiagen, USA). HCV RNA load was determined by Geno-Sen’s HCV real-time PCR Kit (Professional Biotech Pvt. Ltd, India) according to the manufacturer’s instructions using Rotor Gene (Corbett Research, Australia). The real-time PCR program comprised 15 minutes at 50°C (for cDNA synthesis), 10 minutes at 95°C (for initial denaturation) for 45 cycles, including denaturation of 95°C for 15 seconds, 20 seconds annealing at 55°C and an extension for 15 seconds at 72°C.
4. Results
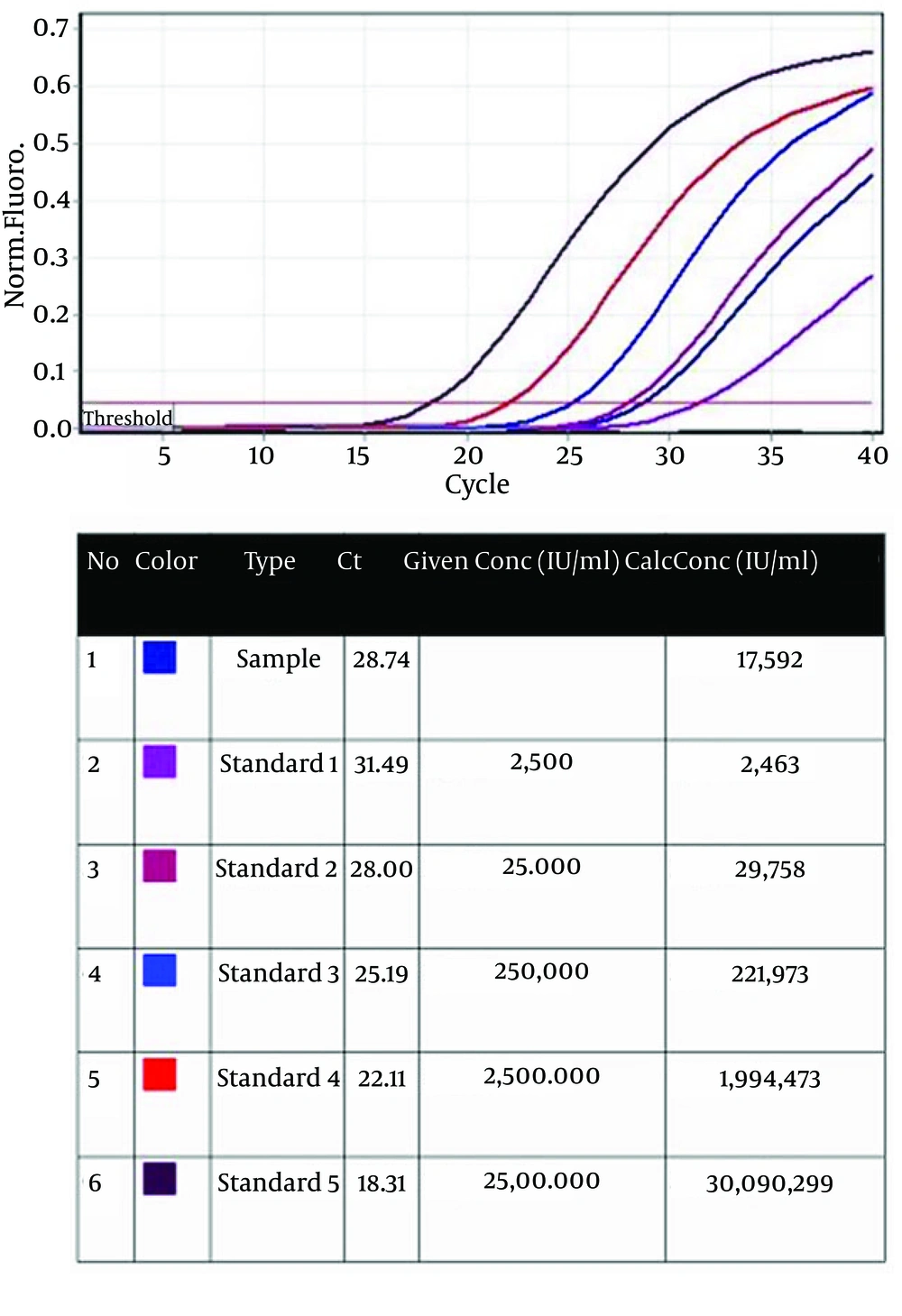
For viral load analysis, viral RNA was purified form collected supernatant of re-infected Huh-7.5 cells and was measured by real-time PCR. In the final sample, a viral load of 1.76 × 104 IU/mL (r2 = 0.997, Median 1.6 × 104 IU/mL) was detected (Figure 1) compared to the standard concentrations. Sample CT was in conformity with standard 2. This indicates the high accuracy of the test.
Five standards from 2.5 × 103 to 2.5 × 107 IU/mL were used. Type (column 3): standards and sample arrangements; Ct (column 4): Ct value of standards and sample; Given Conc (IU/mL) (column 5): quantification standard concentrations; Calc Conc (IU/mL) (column 6): concentrations of the standards and sample that was measured by Rotor Gene.
5. Discussion
In order to set up a cell culture system, a specific cell line, able to sustain viral replication and a plasmid, containing the virus genome, which can multiply in this cell line are required. HCV can multiply in primary hepatocyte and lymphocyte cells but its replication is not permanent. Huh-7 cells were originated from heptocarcinoma, which originally obtained from a 57-year-old Japanese patient suffering from liver tumor in 1982 (9, 12-16). Although development of the Huh-7 cell line was a great breakthrough in designing a model for studying HCV, these cell lines have some restrictions. Among them, low virus yields and its limited spread are of significance (17).
Other derivatives of this cell line are up to 50 folds more permissive compared to classic Huh-7 cell line because of more spread of the infected viruses. Compared to Huh 7, Huh-7.5 cell line has a point mutation in RIG-1. This is responsible for the increased permissivity of Huh-7.5 to HCV. In fact, replicating form of ssRNA viral genome, dsRNA, of viruses such as HCV can activate RIG-1, which in turn leads to phosphorylation of IFN regulatory factor 3 followed by activation of innate immune responses against the virus. Deficiency in RIG-1 pathway confers resistance to viral multiplication. Also, recent studies showed that high expression levels of CD81 and SR-B or clladin-1 can improve viral multiplication and spread in a cell line (17-21). In a study in 2005, Kato and Wakita (15) reported 2.04 × 103 copies of HCV RNA units per mL. However, in the present study, we obtained HCV RNA levels of 1.76 × 104 IU/mL that is about 8.6 times more than that of reported viral loads in previous studies (9, 15). In other studies, HCV was cultured in vitro and its infectivity was confirmed by neutralization test using anti-CD81 and anti-E2. Also, its genome was detected by RT-PCR, its replication was blocked by interferon, and 50 nm particles were detected by immunoelectron microscopy and density gradient analysis.
Compared to other HCV strains, JFH1 could produce higher titers of virus particles and its production of core antigen was 50 times higher than that of other strains. In addition, JFH1 RNA titer was 1 to 2 logs higher than those of other strains and its infectivity titer was 102 pfu/mL (11, 17, 22). JFH1 causes a typical course of infection in animal models, but recent studies showed that it was attenuated and induced a transient infection in chimpanzee. Therefore, new chimeric strains may be useful for animal model studies. Designing chimeric genomes may provide a new way to increase viral production in cell cultures. However, there is genetic incompatibility between JFH1 and other genomic segments of other genotypes of HCV, which is replaced the same fragment in JFH1 genome. This obstacle can be resolved by using a serial passage of designed chimeras in cell cultures. This procedure leads to the accumulation of adaptive mutations, which increases virus yields (22, 23).
Indeed, cell culture condition may affect viral production. Many studies showed that certain mutations mainly in structural proteins may be important for viral release and final virus production. In some studies, chimeric HCV were constructed based on JFH1 genomes. In these chimeric viruses, JFH1 core to NS2 regions were replaced with those genes of other genotypes. These chimeric viruses replicate efficiently in Huh-7 cells and produce infectious viral particles both in tissue cultures and animal models. There are different methods to quantify viral production. Plaque assays, TCID50, fluorescent focus assay, and protein assay are classic methods that are generally slow, time consuming, and with low sensitivity. Modern methods such as flow cytometry, quantitative polymerase chain reaction (qPCR), and enzyme-linked immunosorbent assay (ELISA) are now used with more sensitivity and less time-consuming. In the present study, we used real-time PCR for the quantification of viral load. This method cannot detect whole viral particles but can measure relatively low copy number of viral genomes. Therefore, real-time PCR is more sensitive in comparison to other methods for the measurement of viral loads (24-26).
In the present study, a cell culture system for HCV was developed and titer of the propagated virus was measured by real-time PCR. The result of real time PCR showed a high titer of the virus. Establishment of this cell culture system can support future studies on new antiviral agents and vaccines against HCV. In addition, it may help us in designing new chimeric viruses for our further studies.