1. Background
Coagulase-negative staphylococci (CNS) are a common cause of nosocomial infections. Because these organisms are a normal commensal on the skin and the differentiation of the infection from colonization and contamination may be difficult, the epidemiological studies of CNS infection is complicated (1). The types of CNS infections in children are broad and well described. Pediatric oncology patients, bone marrow transplant recipients, and children with burns are at high risk for acquiring CNS infections (2). The most frequently encountered CNS species associated with human infections is Staphylococcus epidermidis, especially in association with intravascular catheters. Other CNS species such as S. saprophyticus are involved in a variety of infections. This bacterium is an important pathogen in human urinary tract infections, especially in young females (3).
The development of antibiotic-resistant bacteria has increased at a frightening rate since the introduction of antibiotics in the 1940s. The frequent overuse of antibiotics, incorrect diagnoses, inappropriate prescribing, non-compliance with antibiotic therapy by patients, and the misuse of antibiotics have all promoted the rapid spread of resistance even to modern antibacterial agents (4). Methicillin-resistance in staphylococci is mediated by the mecA gene, which encodes for the penicillin-binding protein 2A (PBP2A), resulting in decreased affinity for the β-lactam antibiotics (5). This gene is widely disseminated among Staphylococcus species, which might be due to horizontal transmissions among CNS isolates and S. aureus (6, 7). Serious nosocomial infections caused by methicillin-resistant staphylococci (MRS) require the administration of non-β-lactam antibiotics. An increase in MRS in recent years has led to the severity of the disease caused. The high cost of antibiotics and increasing resistance to them justify the identification, prevention, and control of these infections.
2. Objectives
The purpose of this study was to evaluate methicillin-resistant coagulase-negative staphylococci (MRCNS) isolates with three methods, consisting of oxacillin disk agar diffusion, oxacillin agar screening, and polymerase chain reaction (PCR). Also, the surveillance of the antibacterial resistance patterns of our isolates was our next objective.
3. Patients and Methods
3.1. Sample Collection
In the present study, 122 CNS isolates were obtained from various clinical specimens, including the blood, urine, wound, and body fluids. These samples were collected at four different centers in Iran, including Pediatric hospital of Tehran (No. 1), Imam Reza hospital (No. 2), Aalinasab hospital of Tabriz (No. 3), and Fatemiyyeh hospital of Shabestar (No. 4) from July 2012 to December 2013.
3.2. Isolation and Identification of Coagulase-Negative Staphylococci
In this study, all of the isolates were identified as CNS by different biochemical tests such as Gram staining, catalase, coagulase tube test, ability to growth on mannitol salt agar, deoxyribonuclease (DNase), and novobiocin resistance test (8). Staphylococcus aureus ATCC 29213 (oxacillin susceptible) and S. aureus ATCC 33591 (oxacillin resistant) were used as negative and positive controls, respectively.
3.3. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility of CNS and the reference strains (S. aureus ATCC 29213 and ATCC 33591) were determined by the conventional disk agar diffusion test according to The Clinical and Laboratory Standards Institute (CLSI) recommendations (9). Briefly, disks of different antibiotics (Mast, UK), including oxacillin (1 μg/disk), penicillin (5 μg/disk), erythromycin (15 μg/disk), clindamycin (2 μg/disk), cefazolin (30 μg/disk), rifampicin (5 μg/disk), ceftriaxone (30 μg/disk), sulfamethoxazole/trimethoprim (1.25 - 23.75 μg/disk), tetracycline (30 μg/disk), fusidic acid (10 μg/disk), ciprofloxacin (30 μg/disk), vancomycin (30 μg/disk), teicoplanin (30 μg/disk), amikacin (30 μg/disk), and gentamicin (10 μg/disk), were placed on the plates and incubated for 24 hours and examined for inhibition zones.
3.4. Oxacillin Agar Screening
All the isolates were cultured on Mueller-Hinton agar (Pronadisa, Spain) supplemented with 4% (wt/vol) NaCl comprising oxacillin (Sigma, Australian) at concentrations of 0.6, 4, or 6 μg/mL. The agar plates were inoculated by the swabbing of the surface with adjusted inoculums suspensions (0.5 McFarland) (9). Oxacillin resistance was confirmed by bacterial growth after 24 and 48 hours of incubation at 35°C.
3.5. DNA Extraction
The genomic DNA of the total isolates was extracted via the Cetyltrimethylammonium Bromide (CTAB) method (10) and stored at 20ºC. A loopful of bacteria was added to sterile distilled water (1.5 ml), mixed gently, and was centrifuged in 1000 g for 10 minutes. The supernatant was discarded and Tris-EDTA buffer (TE buffer) (270 μL) plus sodium dodecyl sulfate (SDS) 10% (30 μL) plus proteinase K (5 μL) was added to a microtube and then incubated for 1 hour at 65ºC. Afterward, M NaCl solution (100 μL) was added to the microtube and mixed well. Then, prewarmed CTAB/NaCl (65ºC) solution (80 μL) was added to the microtube and incubated at 65ºC for 10 minutes. Chloroform-isoamylalcohol (24 : 1) solution (700 μL) was added to the microtube and vortexed for 20 seconds. The suspension was centrifuged in 12,000 g for 5 - 10 minutes at 10ºC and the aqueous phase was transferred into a new microtube. Thereafter, isopropanol (200 - 300 μL) was added to each microtube and mixed gently and incubated at 20ºC for 30 minutes before it was finally centrifuged in 12,000 g for 10 minutes. The supernatant was discarded, and the pellet was resuspended in 70% cold ethanol (1 ml) and then centrifuged in 12,000 g for 5 minutes at 10ºC. The supernatant was discarded and after air drying, the DNA pellet was dissolved in TE (10 : 1) buffer (50 μL) and incubated at 37ºC for 30 minutes before it was stored at 4ºC overnight.
3.6. Detection of the mecA Gene by Polymerase Chain Reaction Method
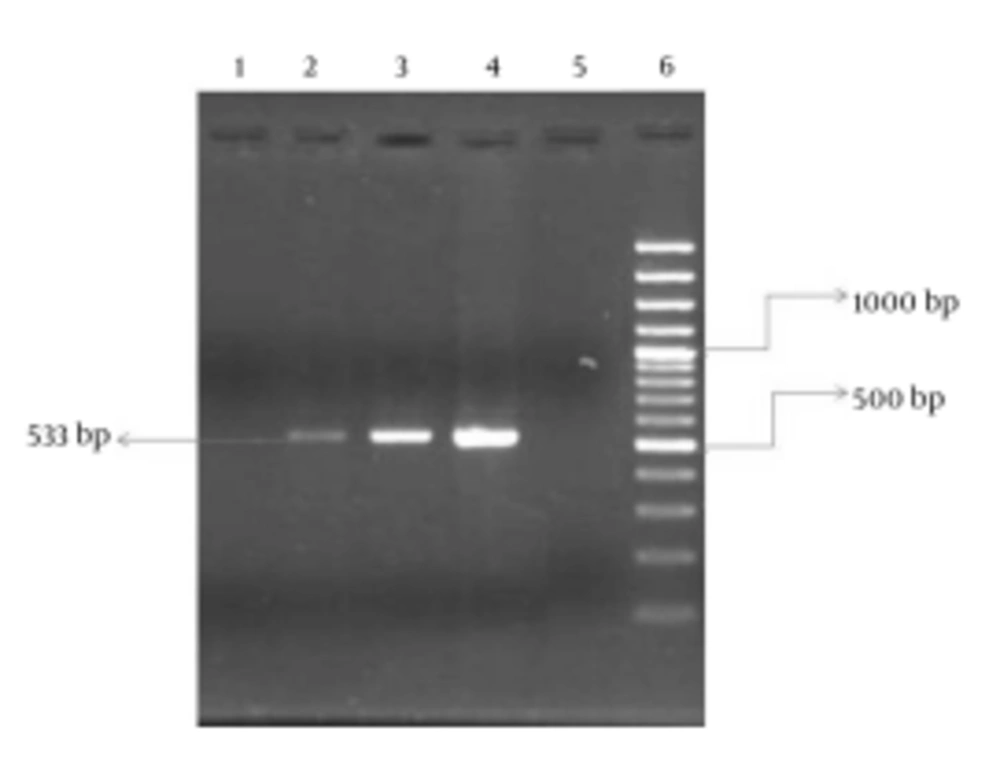
A partial mecA gene was amplified using mecA gene primers (mecA-F: 5’-AAAATCGATGGTAAAGGTTGGC-3’ and mecA-R: 5’-AGTTCTGCAGTACCGGATTTGC-3’), which were selected on the basis of the published nucleotide sequence (11, 12). The PCR reaction mixture was prepared in a reaction mixture (50 µL), containing Tris-HCl (10 mmol/L), 0.2 mmol/L of each deoxynucleotide, MgCl2 (1.5 mmol/L), 25 pmol of each primer, and Taq polymerase (2.5 units) (CinnaGen, Iran). PCR amplification consisted of one cycle at 94°C for 3 minutes, followed by 30 cycles at 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 30 seconds, and then finally 72°C for 3 minutes. For the analysis of the mecA gene, primers mecA-F and mecA-R yielded a fragment of 533 bp. Therefore, the PCR products were examined using 2% agarose gel (Kawsar Biotech Company, Iran) electrophoresis. The gels were stained with ethidium bromide (Geneon Company, Germany) to detect of the fragment of the mecA gene. Also, the 100 bp plus DNA ladder (Fermentas Company, Iran) was used as a DNA molecular weight standard.
4. Results
In this study, 122 CNS strains (110 S. epidermidis and 12 S. saprophyticus isolates) were isolated from various samples, including 66 (54.1%), 41 (33.6%), 12 (9.8%), and 3 (2.5%) isolates from blood, urine, body fluid, and wound specimens, respectively. Also among the 122 CNS, 66 strains were separated from females and 56 strains from males. Ninety-three (76.3%) CNS isolates were resistant to oxacillin (MRCNS) according to disk agar diffusion. Also, oxacillin agar screening using three concentrations was performed for all the isolates, the results of which are shown in Table 1. In our study, the presence of the mecA gene was investigated in 92 (74.5%) of the isolates by PCR (Figure 1).
aData are presented as %.
Our results demonstrated that simultaneous resistance to several antibiotics in the mecA-negative isolates was less than that of the mecA-positive isolates. In other words, the mecA-negative isolates had a high level of susceptibility to all the antibiotics other than penicillin. The results of our study also showed that all the CNS were susceptible to vancomycin and no strains resistant to this antibiotic were observed by disk agar diffusion. For example, according to the comparison of antibiotic resistance patterns between the mecA-positive and mecA-negative isolates, as is depicted in Table 2, 96.3% of the mecA-negative isolates and 100% of the mecA-positive isolates were resistant to penicillin. In this study, 10 (8.2%) outpatient isolates and 82 (67.2%) patient isolates revealed mecA gene, which was statistically significant (P < 0.05). Information regarding the source of the isolates and their resistance patterns is presented in Table 3. Although 90% of the isolates were recovered from Aalinasab hospital, no significant association was detected between the strains isolated from the different hospitals and this gene.
| Antimicrobial Agents | mecA-Positive (n = 92) a | mecA-Negative (n = 30) a |
|---|---|---|
| Oxacillin | 83 (90.2) | 10 (33.4) |
| Penicillin | 92 (100) | 22 (73.3) |
| Rifampicin | 17 (18.5) | 2 (6.7) |
| Erythromycin | 78 (84.8) | 15 (50) |
| Clindamycin | 53 (57.6) | 7 (23.2) |
| Cefazolin | 32 (34.7) | 1 (3.3) |
| Ceftriaxone | 75 (81.5) | 10 (33.3) |
| Co-trimoxazole | 67 (72.8) | 13 (43.4) |
| Gentamicin | 50 (54.4) | 2 (6.6) |
| Ciprofloxacin | 44 (48.9) | 5 (16.6) |
| Vancomycin | 0 (0) | 0 (0) |
| Fusidic acid | 23 (25) | 6 (20) |
| Tetracycline | 56 (60.9) | 13 (43.4) |
| Amikacin | 24 (26.1) | 2 (6.6) |
| Teicoplanin | 7 (7.6) | 0 (0) |
a Values are presented as No (%).
| Antimicrobial Agents | Hospitals | |||
|---|---|---|---|---|
| No. 1 (n = 61) | No. 2 (n = 19) | No. 3 (n = 20) | No. 4 (n = 22) | |
| Oxacillin | 44 (72.1) | 14 (73.7) | 19 (95) | 16 (72.7) |
| Penicillin | 59 (96.7) | 16 (84.2) | 20 (100) | 19 (86.4) |
| Rifampicin | 8 (13.1) | 3 (15.8) | 5 (25) | 3 (13.6) |
| Erythromycin | 49 (80.3) | 12 (63.1) | 19 (95) | 14 (63.6) |
| Clindamycin | 32 (52.4) | 7 (36.8) | 14 (70) | 7 (31.8) |
| Cefazolin | 17 (27.8) | 3 (15.8) | 8 (40) | 6 (27.3) |
| Ceftriaxone | 47 (77) | 11 (57.9) | 14 (70) | 13 (59.1) |
| Co-trimoxazole | 38 (52.3) | 14 (73.7) | 16 (80) | 12 (54.5) |
| Gentamicin | 28 (45.9) | 10 (52.6) | 10 (50) | 4 (18.2) |
| Ciprofloxacin | 20 (32.8) | 9 (47.4) | 14 (70) | 7 (31.8) |
| Vancomycin | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Fusidic acid | 10 (16.4) | 6 (31.6) | 4 (20) | 9 (40.1) |
| Tetracycline | 30 (49.2) | 11 (57.9) | 16 (80) | 12 (54.5) |
| Amikacin | 15 (24.6) | 4 (21) | 5 (25) | 2 (9.1) |
| Teicoplanin | 6 (9.8) | 0 (0) | 1 (5) | 0 (0) |
a Values are presented as No (%).
5. Discussion
Numerous studies have been conducted to determine the optimal methods for the phenotypic detection of methicillin resistance in CNS (13, 14). In this study, we compared two phenotyping methods and genotyping methods to evaluate the oxacillin susceptibilities of CNS. We found that disk agar diffusion showed the lowest sensitivity and specificity (89.2% and 69%, respectively) for the detection of oxacillin resistance between the two methods (Table 1) inasmuch as 9 isolates that contained the mecA gene were found to be susceptible to oxacillin and 10 isolates that did not contain the mecA gene were found to be resistant to oxacillin by disk diffusion method. This finding may be associated with heteroresistance to oxacillin and the absence and presence of the mecA gene expression in these isolates (15, 16). The detection of oxacillin resistance among CNS isolates is difficult mainly because it is often heterogeneous. Over the years, various methods have been employed to overcome this particular obstacle (15).
The use of agar screening at a concentration of 6 µg/mL of oxacillin and 24 - 48 hours of incubation is no longer recommended by the CLSI. However, many studies have observed that this technique is sensitive and can be used as an additional test to acknowledge the results obtained by disk agar diffusion and PCR (14, 17). In our study, the results obtained by this method showed a good relation with those obtained by PCR (93.7% sensitivity and 88.9% specificity). Our results showed that agar screening at a concentration of 4 µg/ml and 0.6 µg/mL of oxacillin and 24 - 48 hours of incubation presented 94% sensitivity and 100% specificity. Not only is this test the most accurate method for the detection of oxacillin resistance among CNS isolates but also it is easy to perform and cheap, which makes it a suitable alternative to PCR.
Agar screening at concentrations of 6, 4, and 0.6 µg/mL of oxacillin and 24 - 48 hours of incubation, as is described here, conferred a reliable detection of the mecA-positive and mecA-negative CNS clinical isolates. The accurate diagnosis of MRCNS strains in hospitals, patients, and health care workers is an important need, and continuous surveillance of antibiotic resistance patterns among CNS strains should receive due attention from health care systems (18). Antibiotic-resistant CNS has emerged as a major cause of morbidity and mortality in the hospital setting during the last decade. Many studies from different parts of the world have reported the presence of multidrug resistance in CNS (19). A comparison between the antibiotic sensitivity patterns of the MRCNS and the methicillin-susceptible coagulase-negative staphylococci (MSCNS) in our study revealed that the MRCNS had a higher level of resistance to many antibiotics than the MSCNS. Many authors have reported similar findings showing higher antibiotic resistance among MRCNS isolates (12, 18, 20).
The results of our study indicated that the three most effective drugs against the MRCNS were vancomycin, teicoplanin, and rifampicin. Vancomycin is the last resort and drug of choice to treat infections caused by MRCNS isolates in the world; therefore, the emergence of resistance to vancomycin could be a serious concern for public health (21, 22). Fortunately, vancomycin exhibited 100% effectiveness, which chimes in with the results of previous studies (22, 23). During the last decade, S. aureus and CNS have emerged as important nosocomial pathogens, and the rising antibiotic resistance in these organisms is a major public health concern (23, 24). The recognition and discrimination of S. aureus and CNS and the detection of methicillin resistance are essential for prompting effective antimicrobial therapy and for limiting the unnecessary use of certain antibiotics (25). Our results revealed that the MRCNS and MSCNS isolates were most resistant to penicillin, with a resistance frequency of 100% and 73.3%, respectively. This finding is in agreement with other reports from Iran and other countries (26-28). We observed that the resistance rate to different antibiotics among the MRCNS strains was higher than those sensitive to methicillin, which is consistent with the results of another study (29). In addition, multidrug resistance rates in our MRCNS and MSCNS isolates were 93% and 56%, respectively. Other published reports have indicated a closely similar or lower percentage of resistance (30-32).
The majority of the strains isolated from the Pediatric hospital of Tehran and Aalinasab hospital of Tabriz were MRCNS, while the number of the MRCNS and MSCNS isolated from the other hospitals was almost equal. These findings were statistically significant (P ˂ 0.05). MRCNS are emerging nosocomial pathogens and every effort should be made to control and prevent infections. Effective infection control programs should be devised and the risk factors should be minimized through regular surveys of health care providers to detect and treat the CNS carriers (18). The current study showed that 76.2% of the CNS isolated in Iran hospitals during a 6-month period were resistant to methicillin. The results also revealed an increase in the number of MRCNS with reduced susceptibility to fusidic acid, rifampicin, and teicoplanin.