1. Background
Escherichia coli is one of the important members of Enterobacteriaceae, which causes some of the common bacterial infections such as urinary tract, bacteremia and traveler’s disease (1). Because of colonization in the intestine, E. coli is considered as enteric bacteria (2, 3). Chromosomal mutation is one of the mechanisms of antibiotic resistance that occurs in the absence of antibiotic, yet the secondary resistance mechanism, is genetic interchanges that is mainly conducted by plasmids. This kind of resistance in bacteria has also been reported against sulfonamides (4, 5).
Sulfonamides are structural analogues of para amino benzoic acid (PABA), which competitively inhibit dihydropteroate synthetase activity. Simultaneous prescription of dihydrofolate reductase (trimethoprim) with sulfonamides creates a synergistic antimicrobial activity in bacterial infections. Combination of trimethoprim and sulfamethoxazole with the trade name of cotrimoxazole is the first antibiotic that has been used for the treatment of urinary tract infections (5, 6).
Dihydropteroate synthetase with low affinity for sulfonamides is encoded on a plasmid, which has high-speed transfer potential to other organisms. Furthermore, sul1, sul2, and sul3 are known plasmid encoded sulfonamide resistance genes, which produce dihydropteroate synthetase (DHPs) and induce resistance against sulfonamides (6, 7). High prevalence of sulfonamide resistance between Gram-negative bacteria, including E. coli isolated from human and animal resources, have been reported worldwide (8-10).
2. Objectives
Because of increasing antibiotic consumption and in turn antibiotic resistance, determination of antibiotic susceptibility patterns of E. coli against cotrimoxazole and study of related genes are necessary for elucidating molecular and epidemiological mechanisms of resistance.
3. Materials and Methods
3.1. Bacterial Isolates
One-hundred and forty-four (144) clinical strains of E. coli were collected from hospitals of Ilam and the Milad Hospital of Tehran. All of the antibiotic discs used in this study were from Mast, England. Antibiotic susceptibility for ampicillin, tetracycline, ciprofloxacin and cotrimoxazole was done by the disk diffusion method based on recommendations of the national clinical laboratory standards (NCLS) (11, 12).
3.2. Phenotypic Detection of Extended Spectrum Beta Lactamase
Extended spectrum beta lactamase (ESBL) producers were confirmed by a disc diffusion method. The pairs of discs tested included cefotaxime-clavulanic acid (30/10 µg) and cefotaxime (30 µg) discs, and ceftazidime-clavulanic acid (30/10 µg) and ceftazidime (30 µg) discs. An increase in the zone diameter of ≥ 5 mm for the clavulanic acid-supplemented discs compared to the zone diameter for the plain discs was considered as an indication of an ESBL-producer. Culture media, agarose and ethidium bromide used in the study were purchased from Merck, US. Escherichia coli ATCC 700603 was used as the ESBL-positive control (13).
3.3. DNA Extraction and Polymerase Chain Reaction Studies
Cotrimoxazole resistance genes sul1, sul2, sul3, dfrA1, dfrA5, int1, blaTEM, blaSHV and CTX-M were detected by PCR. The presence of class 1 integrons (int1) in each strain was assessed by using class 1 specific primers. Extended Spectrum Beta Lactamase genes blaTEM, blaSHV and CTX-M were detected by PCR. A fresh bacterial colony was suspended in 100 mL of sterile distilled water and boiled at 100°C for 10 minutes. After centrifugation, 3 µL of supernatant was used for PCR assays with the primers described in Table 1. Amplification of DNA was performed in a thermal cycler (Eppendrof, Germany). All of the enzymes of the study were purchased from Thermo, US. Polymerase Chain Reaction elongation times and temperature conditions are described in Table 1. The PCR products were electrophoresed on 1.5% agarose gel and visualized under UV light (Biorad, US) (14).
| Gene | Primer | Size, bp | Annealing Temperature, °C | Reference |
|---|---|---|---|---|
| sul1 | 432 | 55 | Present study | |
| F: 5’-CGGCGTGGGCTACCTGAACG-3’ | ||||
| R: 5’-GCCGATCGCGTGAAGTTCCG-3’ | ||||
| sul2 | 293 | 53 | Present study | |
| F: 5’-GCGCTCAAGGCAGATGGCATT-3’ | ||||
| R: 5’-GCGTTTGATACCGGCACCCGT-3’ | ||||
| sul3 | 569 | 55 | Present study | |
| F: 5-CAGATAAGGCAATTGAGCATGCTCTGC-3 | ||||
| R: 5–GATTTCCGTGACACTGCAATCATT-3? | ||||
| dfrA5 | 279 | 53 | (15) | |
| F: 5-ACGGAGTGATTGGTTGCGG-3 | ||||
| R:5-CTCTGTAAATCTCCCCGCC-3 | ||||
| dfrA1 | 334 | Present study | ||
| F: 5-TGGAGTTATCGGGAATGGC-3 | ||||
| R:5-AACATCACCTTCCGGCTCG-3 | ||||
| int1 | 585 | 56 | (15) | |
| F: 5-GCCTGTTCGGTTCGTAAGCT-3 | ||||
| R: 5-CGGATGTTGCGATTACTTCG-3 | ||||
| blaTEM | 861 | 53 | Present study | |
| F: 5-ATG AGT ATT CAA CAT TTC CGT-3 | ||||
| R: 5-TTA CCA ATG CTT AAT CAG TGA-3 | ||||
| blaSHV | 927 | 53 | Present study | |
| F: 5-GGG TTA TTC TTA TTT GTC GC-3 | ||||
| R: 5-TTA GCG TTG CCA GTG CTC-3 | ||||
| CTX-M | 759 | 55 | Present study | |
| F: 5-ACGCTGTTGTTAGGAAGTG-3 | ||||
| R: 5–TTGAGGCTGGGTGAAGT-3 |
3.4. Determination of Cotrimoxazole Minimum Inhibitory Concentration
Minimum Inhibitory Concentration (MIC) values for cotrimoxazole were determined by broth macrodilution of cotrimoxazole-resistant isolates (n = 30). Only isolates that showed resistance to cotrimoxazole (irrespective of resistance to other antibiotics) were selected for MIC determination. Escherichia coli ATCC 25922 was used as the control strain in both Kirby-Bauer disk diffusion and broth macrodilution tests (12).
3.5. Curing of Plasmids by Ethidium Bromide
Techniques were similar as that described by Ansary (16). The R plasmids were studied for curability in cotrimoxazole resistant E. coli isolates. Nutrient broth (5 mL) tubes containing 100 µg/mL concentration of ethidium bromide were incubated with log phase cultures of E. coli host bearing R plasmids to give a 20-fold dilution. A control tube lacking ethidium bromide was always included. All the tubes were incubated over night at 37°C. The contents of the control tube and of the ethidium bromide-containing tubes were cultured on Muller Hinton Agar (MHA), to obtain isolated colonies, while appropriate dilutions of the culture were inoculated on MHA to obtain single colony isolates. After overnight incubation at 37°C, the resulting colonies were tested for loss of antibiotic resistance on MHA plates containing subMIC concentrations of cotrimoxazole (17).
4. Results
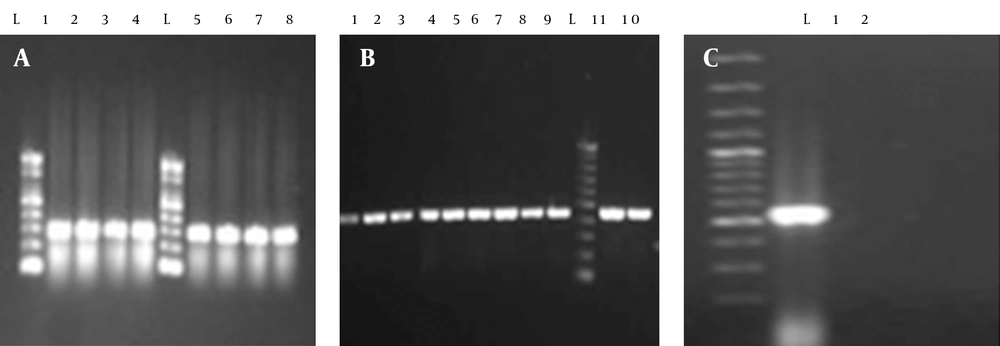
A total of 144 clinical isolates were collected from patients admitted to different wards of the Milad Hospital (Tehran) and various hospitals of Ilam (Imam khomaini and Shahid Mostafa), Iran. Out of 144 strains of E. coli isolated from urinary tract infections 87 (60.41%), 117 (81.25%), 84 (58.33%), 5 (3.47%) and 59 (40.97%) were resistant to cotrimoxazole, ampicillin, tetracycline, nitrofurantoin and ceftriaxone, respectively (Table 2). Out of 87 cotrimoxazole resistant isolates, 71 (81%) had the sul1 gene (Figure 1). Fifty-nine (67%) and two (2.29%) of the strains had the sul2 and sul3 genes, respectively (Figure 1). Forty-four (50.57%) strains had the sul2 and sul1 genes, simultaneously (Figure 1). Two (2.29%) of the isolates possessed all of the studied genes, namely sul1, sul2 and sul3 (Figure 1). Thirty-four (39.1%) and five (5.7%) of the isolates had the dfrA1 and dfrA5 gene, respectively. Sixty-eight (78.2%) strains had the int1 gene. Furthermore, dfrA1 and dfrA5 were present in three (3.4%) of the isolates.
| Cotrimoxazole Resistance Genes | Isolates |
|---|---|
| sul1 | 71 (81.6) |
| sul2 | 58 (66.7) |
| sul3 | 2 (2.3) |
| sul1 + sul2 | 44 (50.6) |
| sul1 + sul3 | 2 (2.3) |
| sul2 + sul3 | 2 (2.3) |
| sul1 + sul2 + sul3 | 2 (2.3) |
| dfrA1 | 34 (39.1) |
| dfrA5 | 5 (5.7) |
| int1 | 68 (78.2) |
| dfrA1 + dfrA5 | 3 (3.4) |
| dfrA1 + int1 | 27 (30.3) |
| int1 + dfrA5 | 5 (5.7) |
| dfrA1 + dfrA5 + int1 | 3 (3.4) |
| sul1 + sul2 + int1 | 36 (41.1) |
| sul1 + int1 | 25 (28.7) |
| sul1 + int1 + dfrA1 | 7 (8.0) |
| sul1 + dfrA1 | 26 (29.9) |
| sul2 + int1 | 42 (48.3) |
| sul2 + dfrA1 | 26 (29.9) |
| sul2 + int1 + dfrA1 | 22 (25.3) |
| int1 + sul1 + sul2 + dfrA1 + dfrA5 | 1 (1.1) |
| sul1 + sul2 + sul3 + dfrA1 + dfrA5 + int1 | 0 (0) |
| sul3 + int1 | 2 (2.3) |
| sul3 + dfrA1 | 1 (1.1) |
| sul3 + dfrA5 | 0 (0) |
| sul3 + int1 + dfrA1 | 1 (1.1) |
| sul3 + dfrA1 + dfrA5 + int1 | 0 (0) |
a Data are presented as No. (%).
4.1. Polymerase Chain Reaction for the Detection of blaTEM, blaSHV and CTX-M Genes
Among the 144 E. coli isolates, seventy-two (50%) ESBL producing and seventy-two (50%) non-ESBL producing E. coli isolates were identified. All of the ESBL-producing E. coli isolates were tested for detection of blaTEM, blaSHV and CTX-M. Our results showed that 85.2% (n = 76), 53.2% (n = 48) and 26.1% (n = 24) of the tested E. coli were positive for blaTEM, blaSHV and CTX-M, respectively. Furthermore, 21.4% (n = 19), 16% (n = 14) and 9% (n = 8) of the isolates had the following gene combinations blaTEM and blaSHV, blaTEM and CTX-M, and blaSHV and CTX-M, respectively. Five percent (n = 6) of the isolates had three genes, simultaneously.
4.2. Plasmid Mediated Resistance
The MIC of 30 cotrimoxazole resistant isolates by broth dilution was 700 µg/mL. Curing of plasmids by ethidium bromide in 30 isolates caused disability of bacterial growth on nutrient agar containing sub-MIC concentrations of cotrimoxazole.
5. Discussion
This is the first report of sul genes from Iranian E. coli isolates. The overall resistance percentage of isolates against trimethoprim/sulfamethoxazole, ampicillin, tetracycline and ciprofloxacin was very high, and the highest resistance was estimated against ampicillin (81.25%), followed by cotrimoxazole (60.41%). Based on our results the sul1 gene has the highest prevalence in E. coli strains resistant to cotrimoxazole. Frequency of sul1 (81%) was higher than sul2 (67%) and sul3 (2.29%), which is in accordance with other studies conducted worldwide (9, 10). Our observed trends in sulfamethoxazole-resistant allele distributions (sul1 > sul3 > sul2) were different from previous studies (9, 15, 18, 19). There have been a few studies of the genetic distributions underlying trimethoprim resistance in the world. Our results agree with four European studies, which have found dfrA1 to be very common in Europe (20, 21). Conversely, studies performed in Korea and Australia found dfrA17 and dfrA12 to be the most common alleles (21-23). In this study prevalence of dfrA1 was more than dfrA5.
Existence of sul genes in different kinds of clinical and environmental isolates indicates that these genes have a universal function of carrying and spreading sulfonamide resistance in bacteria (6, 18, 24-26). For example in a study of 100 environmental E. coli strains, it was revealed that sul1 (50%) and sul2 (60%) genes were present in these isolates at a high percentage (18). The sul1 gene is part of class I integrons in many sulfonamide resistant bacteria (25, 27-30). Class 1 integrons play an important role in antibiotic resistance dissemination in many Multidrug Resistant (MDR) Gram-negative bacteria, including many zoonotic serovars of Salmonella enteric and other enterobacteriaceae (27). We observed regional variation in integron distributions. Prevalence of int1 was positive in 78% (68) of isolates; coexistence of dfrA1 and dfrA5 with int1 was found in 30% and 5.7% of the isolates, respectively. This indicates a correlation between Int1 and dfr1.
Other genetic mobile elements have also been found to act as sources of sul genes. For example, in SMX resistant Vibrio cholerae serogroup 0139, it was reported that the sul2 gene was part of a cluster located on a newly discovered genetic element of the integrative conjugative element group named SXT. The resistance genes of SXT exist in a composite transposon-like structure and were probably acquired recently (30). It is important to mention that frequency and distribution of sul genes and trimethoprim/sulfamethoxazole resistance in various bacteria indicates that these genes are transferred mainly by rapid methods including horizontal routes between E. coli and other bacteria. Many studies have referred to this point (27, 29).
Resistance to cotrimoxazole was observed in ESBL-producing E. coli more than non-ESBL producing E. coli isolates. Frequency of blaTEM was higher than blaSHV and CTX-M. Coexistence of ESBL genes with sul genes in some of the strains probably indicated that these genes are present on one plasmid. Escherichia coli isolates with both sul1 and sul2 were found in ESBL-producing, and non-ESBL producing E. coli isolates. The majority of resistance genes in Gram-negative bacteria exist on cytoplasmic plasmids that spread antibiotic resistance in epidemics. Our results indicated that all of the resistant isolates had sul genes and lost cotrimoxazole resistance after plasmid curing. Sulfonamides are broad-spectrum antimicrobials against Gram-negative and Gram-positive organisms, and have bacteriostatic activities (31).
High prevalence of sulfonamide resistance has been observed in Gram-negatives isolated from human and animals, worldwide (32). According to previous studies, sul1 and sul2 genes equally exist in sulfonamide resistant isolates. However, in Denmark, it was revealed that the prevalence of sul2 is higher than sul1 gene in E. coli strains isolated from humans (24). Grape et al. (25), in their study on sul3 gene in E. coli strains isolated from human resources, reported that out of 64 sulfonamide resistant isolates, 39 and 48 isolates carried sul1 and sul2 genes, respectively. In contrast, 25 isolate simultaneously had sul1 and sul2 genes, while two strains were lacking both of these genes (25). The sul2 gene has been observed with high prevalence in E. coli strains isolated from pig, poultry, cows, human feces and urinary tract infections (6). According to the studies of Wu et al. (7), sul3 gene is least prevalent in E. coli isolates from human and animal resources. Out of 501 isolates, 109 cases were resistant to sulfonamides. Relative prevalence of sul2, sul1 and sul3 genes in this study were 65%, 45% and 12%, respectively, which is different with our results. However, co-existence of class I integrons also existing in plasmids carrying sul1, sul2 and sul3 genes was found in 80%, 100% and 5% of isolates, respectively; this finding is similar with our results, yet the prevalence of this coexistence in our study was lower than that of Wu et al. (7).
Medina et al. (33) in their study of the resistance pattern of more than 400 pathogenic E. coli from ruminants reported very high percentage of resistance, and in their studied strains resistance rate was 76.3% (dhfrI), 60% (sul1) and 63.3% (sul2) for trimethoprim and sulfamethoxazole. Al-Agamy (34) in a comprehensive study of susceptibility patterns of 100 uropathogenic E. coli isolates showed that resistance rates for ampicillin, tetracycline, trimethoprim, sulfamethoxazole/trimethoprim and ciprofloxacin, were 90%, 85%, 70%, 62% and 24%, respectively. The prevalence of the antibiotic resistance genes for sul1, sul2 and sul3 were 68.18%, 86.36% and 5.5%, respectively. The sul2 was the most prevalent resistance gene. The prevalence of class 1 integron was reported to be 95.45% in this study (34). Infante et al. (26) in their study of sulfonamide resistance genes of 20 cotrimoxazole resistant E. coli isolates from children’s feces found that 19 out of 20 strains carried at least one sul gene. Furthermore, sul1 and sul2 genes were detected in 13 isolates, while one isolate carried sul3 and sul2 genes. The sul2 gene was the most prevalent and was observed in 11 strains, while sul1 was detected in four strains (26). In another study of 350 uropathogenic E. coli isolates from Europe and Canada, sul2 (77.9%) was reported as the most common sulfamethoxazole resistant gene in sulfamethoxazole-resistant isolates (28). Recently, numerous studies have been conducted in Iran on cotrimoxazole resistance. Results of such reports indicated that high rate of resistance against cotrimoxazole are present in different genera including E. coli (35).
High frequency of sul genes, plasmid related resistance and high prevalence of SXT resistance in E. coli isolates indicates that continuous surveillance programs should be implemented in hospital and clinical settings to better control and treat related diseases, and monit the trends of SXT resistance in Gram-negative bacteria.