1. Background
Clostridium perfringens, a Gram-positive anaerobic bacterium that produces spores is widely distributed in nature and normally found in the intestines of animals and humans. Based on the four main toxins produced by C. perfringens species (α, β, ε, ι), they are divided into five groups (A, B, C, D and E). In addition to strong metabolic activities, C. perfringens species also produce at least 16 virulence factors which include 12 toxins (α-ν), enterotoxin, hemolysin, and neuraminidase (sialidase). Alpha-toxin or phospholipase C is the leading cause of death and necrosis as the first virulence factor of this microorganism and is produced by all strains of C. perfringens. C. perfringens creates variable pathogenic conditions, ranging from food poisoning to life-threatening myonecrosis (1).
Type A C. perfringens is a major factor distributed in the environment and is the cause of diseases such as Clostridium myonecrosis, food poisoning, necrotic enterocolitis infection in children, necrotic bowel disease (Pig Bel), and antibiotic-associated diarrhea (1, 2). Enterotoxin is produced about 1-5% by type A C. perfringens. The toxin is a single polypeptide chain of about 35 kDa which unlike other toxins is produced and released in the sporulation stage (1, 2). Following food poisoning caused by Salmonella and Staphylococcus, the enterotoxin of C. perfringens is the third leading cause of food-borne diseases worldwide (3). Enterotoxin-producing genes are located on a transposon called Tn5565.
Several studies have shown that the risk of acquiring antibiotic resistance in C. perfringens is high (4). Because of the importance of the treating diseases caused by C. perfringens, extensive researches have been conducted in this area (5, 6). In some of these studies, resistance to the drugs of choice such as penicillin was observed. Resistance to alternative medicines of penicillin like metronidazole and clindamycin has also been observed (5, 6). Despite extensive studies in the world, there are no particular guidelines regarding the management of C. perfringens infections in Tabriz, Iran.
2. Objectives
The aims of this study were culturing and isolation of toxigenic and non-toxigenic C. perfringens, determination of the Minimum Inhibitory Concentration (MIC) for selected antibiotics by the Etest method and molecular study of the enterotoxin gene.
3. Materials and Methods
3.1. Sampling
In a descriptive study, a total of 136 stool specimens, including 56 specimens from hospitalized patients and 40 out-patients as well as 40 samples from healthy individuals of Imam Reza Educational and Medical Center were consecutively collected. Fecal samples of healthy people were collected from people referred to the laboratory for other purposes. These healthy people were justified orally or in some cases were acquired written consent.
3.2. Culturing and Identifying Methods for Clostridium perfringens
At first, cooked meat medium (Que Lab Inc. European division, UK) enrichment method was performed on samples at 45°C. Thereafter, a loopful of the enriched culture was transferred into blood agar and incubated anaerobically (H2 = 10%, N2 = 80%, CO2 = 10%) using Anoxomat and Mart jar at 37°C for 24 - 72 hours. The obtained typical colonies with double zone of hemolysis were identified after conducting the anaerobic tolerance test by different biochemical tests such as Gram staining, phospholipase C (lecithinase) test, indole and urease production, gelatin hydrolysis, and fermentation of sugars like glucose, lactose and mannitol (7).
3.3. The Evaluation of Sensitivity to Antibiotics
MICs for imipenem, metronidazole, ceftriaxone, clindamycin, chloramphenicol, and penicillin were determined by Etests (AB Biomerieux, Sweden). Since in some anaerobic bacteria the growth on Mueller-Hinton agar (Liofilchem Ltd. Italy) was not sufficient, to determine the antibiotic susceptibility of the isolates, enriched brucella agar medium (Fluka Chemie AG CH-9471 Buchs, Switzerland) with vitamin K and hemin were used. All the plates were incubated anaerobically for 24 hours at 37°C and manufacturer’s instructions and Clinical and Laboratory Standards Institute (CLSI) guidelines were considered for testing and reading the obtained results (8).
3.4. Detection of Enterotoxigenic Clostridium perfringens by Duplex Polymerase Chain Reaction
The C. perfringens isolates were cultured on Columbia agar (Himedia Laboratories Pvt. Ltd. India) and the suspension was prepared in distilled water with a net absorption of 0.09-1 at a 600 nm. Afterwards, the suspension was diluted to 1:10 and mixed well. After boiling it for 10 minutes, it was centrifuged with 13000 × g rpm for five minutes and the supernatants were used as templates in the duplex PCR reaction performed with the following specific primers for amplification of 283 bp and 426 bp fragments:
F: ATA GAT ACT CCA TAT CAT CCT GCT
R: AAG TTA CCT TTG CTG CAT AAT CCC (phospholipase C 283 bp)
F: GAA AGA TCT GTA TCT ACA ACT GCT GGT CC
R: GCT GGC TAA GAT TCT ATA TTT TTG TCC AGT (enterotoxin, 426 bp) (9, 10)
The PCR mixture (30 µL) consisted of 3 µL of 10x PCR buffer (the 1x buffer included 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2 and 0.01% gelatin), 0.6 µL of 10 mM dNTP mixture, 1.2 µL (50 pmoles) for CPE and 0.6 µL (25 pmoles) for PLC genes, 1 µL (1 U) of Taq polymerase (Fermentas), 10 µL of the DNA template, and distilled water to make a total volume of 30 µL.
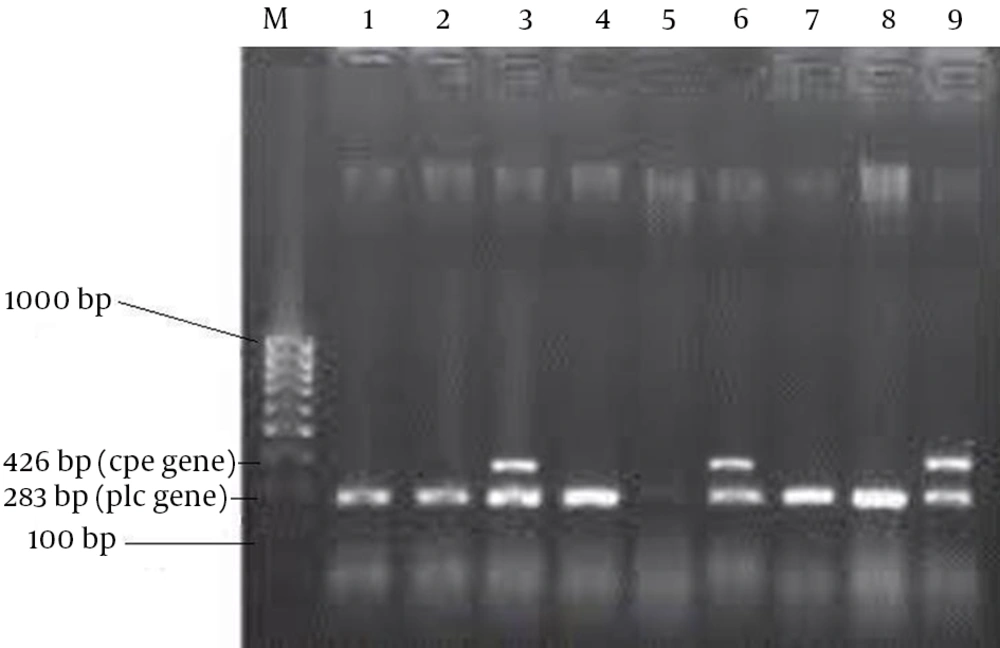
The amplification was performed in a DNA thermal cycler (Eppendorf, Germany), programmed for a primary denaturation, (95°C, 10 minutes), followed by 35 cycles of denaturation (94°C, one minute), annealing (55°C, one minute), elongation (72°C, one minute), and then extension (72°C, 10 minutes). A negative control without template was included in each PCR run (11). The amplified products were visualized by electrophoresis on 2% agarose gel in 1x TBE buffer (1 M Tris, 0.9 M boric acid, 0.01 M EDTA, pH = 8.4), at 80 V, for two hours. A 100-bp DNA ladder was used as a molecular mass marker. The gels were stained with ethidium bromide (0.5 µg mL-1) and photographed on a gel documentation system (UVP, USA) for the analysis of the bands (9-11) (all the PCR materials including primers were provided by CinnaGen; Nedayeh Fan Co., Iran).
4. Results
Of 136 stool samples including diarrhea [48] and non-diarrhea [88] ones, 83 (61.02%) C. perfringens strains were cultured and identified by the above mentioned tests; 33 (68.75%) and 50 (56.81%) of the isolates were obtained from diarrhea and non-diarrhea stool samples, respectively. Of the 83 C. perfringens isolates, 79 showed alpha-toxin (phospholipase C) production gene by PCR; so, were confirmed them as C. perfringens. Totally, 3 (9.09%) and 2 (4.34%) cpe genes were present in diarrhea and non-diarrhea samples, respectively (Figure 1). Of the 79 isolates of C. perfringens, 34 (43.03%) showed no resistance, 18 (22.78%) had one resistance and 27 (34.17%) isolates had multiple resistance to the tested antibiotics. The antimicrobial resistance patterns of the tested isolates are given in Table 1.
| Antibiotics | Antibiotic Content of Etests Strips, µg/mL | Obtained MIC by Etests, µg/mL | CLSI Interpretative MIC, µg/mL | Obtained Results by Etests | |||
|---|---|---|---|---|---|---|---|
| S, % | I, % | R, % | S, No. (%) | R, No. (%) | |||
| Imipenem | 0.002 - 32 | 0.5 - 32 | 4 | 8 | 16 | 49 (62.02) | 30 (37.97) |
| Metronidazole | 0.016 - 256 | 0.2 - 64 | 4 | 16 | 32 | 70 (88.60) | 9 (11.39) |
| Ceftriaxone | 0.016 - 256 | 0.016 - 128 | 16 | 32 | 64 | 77 (97.46) | 2 (2.53) |
| Clindamycin | 0.016 - 256 | ≤ 2 - 32 | 2 | 4 | 8 | 66 (83.54) | 13 (16.46) |
| Chloramphenicol | 0.016 - 256 | 2 - 64 | 8 | 16 | 32 | 75 (94.93) | 4 (5.06) |
| Penicillin | 0.016 - 256 | 0.25 - 4 | 0.5 | 1 | 2 | 72 (91.13) | 7 (8.86) |
aAbbreviations: CLSI, Clinical and Laboratory Standards Institute; I, intermediate; MIC, minimum inhibitory concentration; R, resistant; S, sensitive.
5. Discussion
In cases of serious anaerobic infections, not only should the clinical laboratory be able to accurately identify C. perfringens isolates but it is also important that the laboratory perform susceptibility tests on individual isolates from patients. The precise identification of C. perfringens by different methods is always necessary because identification by biochemical tests in some cases is not satisfactory. In this study, to achieve this goal, in addition to biochemical tests, genes coding production of lecithinase were also detected by PCR, which reduced false positives. Other researchers used commercially available kits such as RapID ANA II, AnIDent, and ATB 32A (12). PRAS biochemical fermentation tests, end product analysis by gas-liquid chromatography and other reactions are also able to confirm the identification of the strains (13, 14).
Clostridia neither invade healthy cells nor multiply within them. They are able to enter the host organisms by two ways, the oral route and wounds, but their proliferation in the intestinal content or in wounds require the presence of risk factors (15). Toxins as the main virulence factors are responsible for all the symptoms and lesions observed in clostridial diseases. Consequently, toxin-producing C. perfringens strains are the main targets for diagnosis of C. perfringens diseases. Enterotoxin-positive strains of C. perfringens are recognized as the causes of food-borne diarrhea as well as several non-food-related diarrheas, including sporadic diarrhea and antibiotic-associated diarrhea (16, 17). In Tabriz, few data are available on the detection of this organism. The analysis of food-borne anaerobic bacteria is not routinely performed due to difficulties in their isolation and identification. The procedures require special techniques which may be expensive, time-consuming and labor intensive. For this reason, the outbreaks of C. perfringens are often not recognized. In addition, the knowledge of antimicrobial susceptibility of this organism isolated from human, animals, foods and environment is limited.
In this study, the enterotoxigenic and non-enterotoxigenic C. perfringens strains were isolated from human feces and identified using biochemical tests and a duplex PCR procedure, which enables C. perfringens species identification and differentiation between enterotoxigenic and non-enterotoxigenic strains by detection of the plc and cpe genes (9, 18). Our results showed that only 79 out of 83 C. perfringens isolates were alpha-toxin (phospholipase C) producers and the presence of the plc gene by PCR confirmed the isolates of C. perfringens (Figure 1). These results indicated that identification only by biochemical tests is not satisfactory. In addition, the MICs of six antimicrobial agents commonly used in human medicine for all the C. perfringens isolated strains were determined using the Etest method. In this study, imipenem was the least active agent to C. perfringens with an overall rate of resistance equal to 37.97%. While this finding is nearly in agreement with another research (19), Camacho et al. reported sensitivity of all his isolates to imipenem (6). Of the three antimicrobials including metronidazole, ceftriaxone and chloramphenicol, which were highly active against C. perfringens with low MIC values, ceftriaxone could inhibit the most C. perfringens strains tested with an overall rate of resistance of 2.53%. These findings are also consisting of Camacho et al. report (6). Penicillin G is still an effective drug based upon its low MIC values and low rate of resistance (8.86%) and may be considered when choosing an antimicrobial agent for prophylaxis or treatment of C. perfringens in humans. In agreement with this, there are several reports (6, 19). The isolates of C. perfringens in this study were susceptible to clindamycin (%83.54), while cases of clindamycin-resistance have been described (20, 21).
This study revealed a moderate prevalence of antimicrobial-resistant C. perfringens strains. Among 56.95% of antimicrobial-resistant strains, 22.78% were single drug resistant and 34.17% were Multidrug Resistant (MDR). This result also has been confirmed by a set of results obtained from Germany (22). We found the presence of the cpe gene among C. perfringens strains isolated from diarrhea stool (9.09%) more than the non-diarrhea samples (4.34%), indicating the role of toxin production in diarrhea (P ≤ 0.05). The C. perfringens strains isolated from various origins carry nearly 6% cpe gene, but this percentage is higher (59%) among the strains isolated from confirmed outbreaks of food poisoning (23). This could be the reason for our results not being in agreement with the results of Tansuphasiri et al. (4.8%) which was much lower than our findings (18).
The fecal samples obtained from the hospitalized patients with non-diarrheal diseases might harbor a small number of these enterotoxigenic strains in their intestines. However, it suggests the need of more studies to evaluate the role of enterotoxigenic C. perfringens in patients with diarrhea as well as non-diarrhea stools. These organisms must be looked for routinely and a periodic evaluation of antimicrobial susceptibility should be performed.