1. Background
Pseudomonas aeruginosa is one of the most important opportunistic pathogens among the sources of nosocomial infections, especially in burns units (1-4). Pseudomonas aeruginosa can grow in hospital environments, which are exposed to heavy antimicrobial use, and can, consequently, be transmitted rapidly among hospitalized burns patients (5). Nosocomial infections caused by P. aeruginosa are often difficult to treat (6). One of the major concerns for the treatment of P. aeruginosa infections is antibiotic resistance since studies have shown that this bacterium has increasingly been resistant to many antibiotics (7, 8). Pseudomonas aeruginosa is naturally resistant to narrow-spectrum Penicillins, first- and second-generation Cephalosporins, Sulfonamides, and Trimethoprim (5). Research has shown that the prevalence of multidrug-resistant P. aeruginosa in burns patients is 87.5% in Iran, 68.8% in Malaysia, and 29.24% in Pakistan (2-4). Carbapenems have been used for the treatment of multidrug-resistant P. aeruginosa infections. However, Carbapenem-resistant isolates have been increasing worldwide and become a major concern for public health (9, 10). Resistance against Carbapenems has emerged as a result of different mechanisms such as impermeability to drugs due to the loss of OprD porin, the upregulation of an active efflux pump system present in this organism, or the production of metallo-beta-lactamases (MBLs), which hydrolyze all Carbapenems (9, 11, 12).
MBLs constitute a group of Ambler class B and have the ability to hydrolyze a wide variety of beta-lactam agents such as Penicillins, Cephalosporins, and Carbapenems. These enzymes require zinc for their catalytic activity and are inhibited by metal chelators such as ethylenediaminetetraacetic acid (EDTA) and thiol-based compounds (13). The genes encoding MBLs are typically part of an integron structure and are carried on large transferable plasmids. The MBL genes can be divided into five categories according to their molecular structures: the IMP; VIM; GIM; SIM; and SPM types (14-17). Molecular typing techniques have become important tools for the provision of information to control cross-infection and to identify the transmission pathways and sources of isolates. In addition, Pulsed-Field Gel Electrophoresis (PFGE) is now the gold standard for DNA fingerprinting techniques because of its high discriminatory power (18).
2. Objectives
We aimed to determine the prevalence and clonal dissemination of MBL-producing P. aeruginosa and its antibiotic susceptibility in the isolates of major hospitals in the western Iranian city of Kermanshah.
3. Materials and Methods
3.1. Bacterial Isolates
Sixty isolates of P. aeruginosa were collected from two hospitals in Kermanshah (Imam Khomeini [A] and Imam Reza [B]) between 2011 and 2012. The specimens were collected from burns (n = 32, 53.3%), urine (n = 9, 15%), respiratory tract secretions (n = 10, 16.7%), and other sources (vagina, catheter, eye, ear, wound, and blood) (n = 9, 15%). The bacteria were then cultured on the MacConkey agar and the Muller-Hinton agar media, and the grown colonies were confirmed using specific tests. These included catalase, oxidase, the growth characteristics and pigment gas production, indole and methyl red, Sulfide Indole Motility, and growth at 42°C on the Cetrimide agar (Merck, Germany).
3.2. Antibiotic Susceptibility Testing
Antibiotic susceptibility testing was performed as recommended by The Clinical and Laboratory Standards Institute (CLSI). Minimal inhibitory concentrations for 4 antibiotics, namely Gentamicin, Piperacillin, Ceftazidime, and Cefepime (MAST, England), were determined using the Micro-Broth Dilution method (19).
3.3. Identification of Metallo-Beta-Lactamases
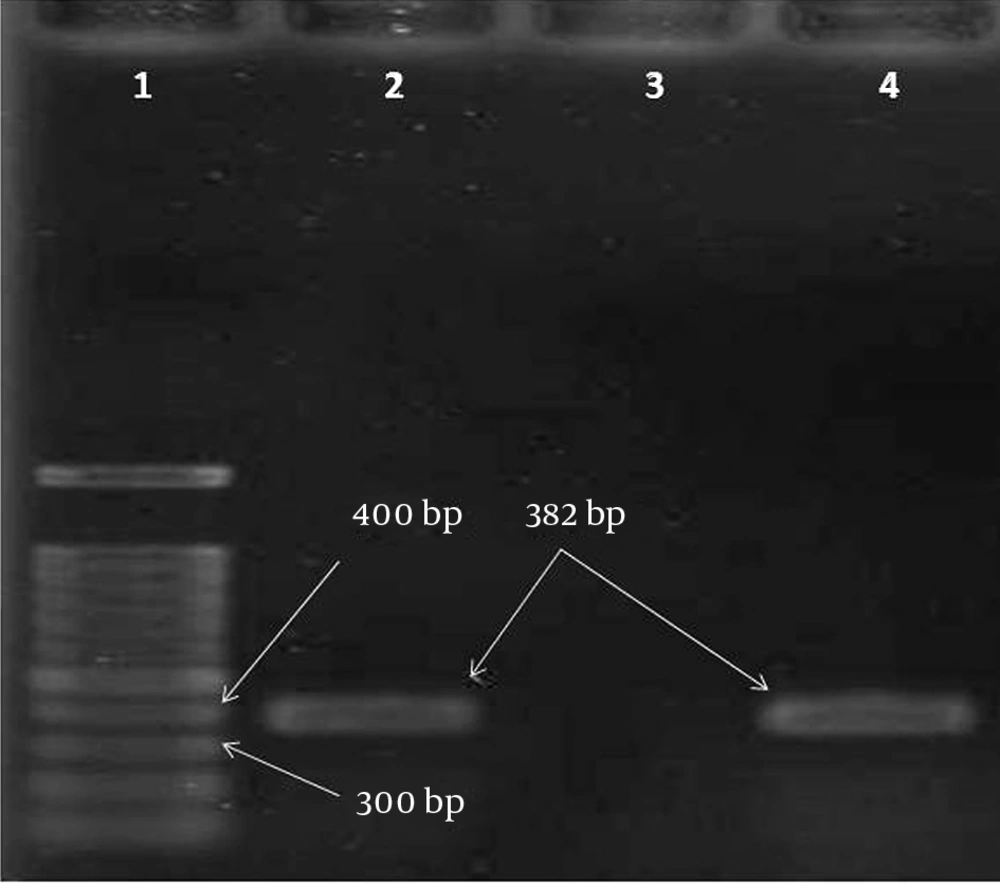
The Double-Disk Synergy Test (DDST) was used to screen the MBL-producing P. aeruginosa isolates (15). Disks containing 930 μg of EDTA (SinaClon, Iran) plus 10 μg of Imipenem (SinaClon, Iran) were placed on the Muller Hinton agar spread by isolates. An increase of ≥ 7 mm in the inhibited zone diameter in the presence of 930 μg of EDTA compared to Imipenem tested alone was considered to be a positive test for the presence of MBLs. For the molecular diagnostic of the MBLs, the boiling method was used for DNA extraction (20). Polymerase Chain Reaction (PCR) was carried out using Master Mix (SinaClon, Iran), DNA, and a pair of primers (SinaClon, Iran). The primers used for the amplification of the VIM were VIMF (5'-GTT TGG TCG CAT ATC GCA AC-3) and VIMR (5-AAT GCG CAG CAC CAG GAT AG-3), which amplified a 382-bp DNA fragment (15). The PCR product was determined using gel electrophoresis (1% agarose) and staining with ethidium bromide, followed by visualization using a Gel Doc apparatus (Bio-Rad, USA).
3.4. Pulsed-Field Gel Electrophoresis
PFGE was performed according to a previously described protocol (21) with some modifications. The MBL-producing P. aeruginosa plugs were digested with 20 U SpeI (Fermentas). Salmonella enterica serovar Braenderup H9812 was digested with 20 U XbaI (Fermentas) and used as an electrophoresis molecular weight marker. Following SpeI digestion, the plugs were loaded into a 1% low-electroendosmosis agarose (Merck, Germany). Electrophoresis was performed on a CHEF Mapper apparatus (Bio-Rad, USA) at 14°C for 22 hours under the following conditions: initial switch time of 5 seconds; final switch time of 35 seconds; included angle of 120°C; voltage gradient of 6 V/cm; and linear ramping factor. The gels were stained with ethidium bromide and visualized under UV light using a Gel Doc apparatus (Bio-Rad, USA).
3.5. Software Analysis
DNA fragment analysis was performed using GelCompar II (version 6.6) (Applied Maths, Belgium). The Dice coefficient was used to calculate similarities, and the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) was employed for cluster analysis. A cluster of isolates was defined to include all isolates with more than 80% similarity in their DNA patterns according to the Tenover criteria (22, 23).
3.6. Statistical Analysis
The statistical analysis of the data was done using the Statistical Package for the Social Sciences (SPSS) (version 16). The chi-square test was utilized to assess the correlation between the data. Statistical significance was defined as a P value < 0.05.
4. Results
4.1. Bacterial Isolates
Of the 60 P. aeruginosa isolates, 47 were from hospital A and 13 from hospital B. The specimens were collected form 31 (51.7%) males and 29 (48.3%) females at an average age of 31.64 ± 23.97 and 28.66 ± 19.90 years old, respectively.
4.2. Antibiotic Susceptibility of Isolates
Of the 60 P. aeruginosa isolates, 30 (50%) were resistant to Gentamicin, 38 (63.3%) to Piperacillin, 42 (70%) to Ceftazidime, and 45 (75%) to Cefepime. The highest resistance to the 4 antibiotics belonged to the isolates from the burns specimens (Table 1).
| Sample Source | Antibiotic Resistance, % | |||
|---|---|---|---|---|
| Cefepime | Ceftazidime | Piperacilin | Gentamicin | |
| Burns | 90.6 | 84.4 | 78.1 | 56.2 |
| Urine | 44.4 | 33.3 | 44.4 | 22.2 |
| Respiratory Tract | 60 | 60 | 40 | 50 |
| Other | 66.7 | 66.7 | 55.6 | 55.6 |
| Total | 75 | 70 | 63.3 | 50 |
4.3. Screening for Metallo-Beta-Lactamase by Double-Disk Synergy Test and Polymerase Chain Reaction
The DDST test determined that 29 (48.3%) isolates were MBL producers. Of this total, 5 (8.3%) strains were positive for the VIM gene (Figure 1) and 4 were from the burns specimens. Of the VIM-producing isolates, 4 (80%) were resistant to Cefepime, Ceftazidime, and Piperacillin. The data analysis via the chi-square test showed no significant association between resistance to antibiotics and MBLs (P = 0.5).
4.4. Pulsed-Field Gel Electrophoresis
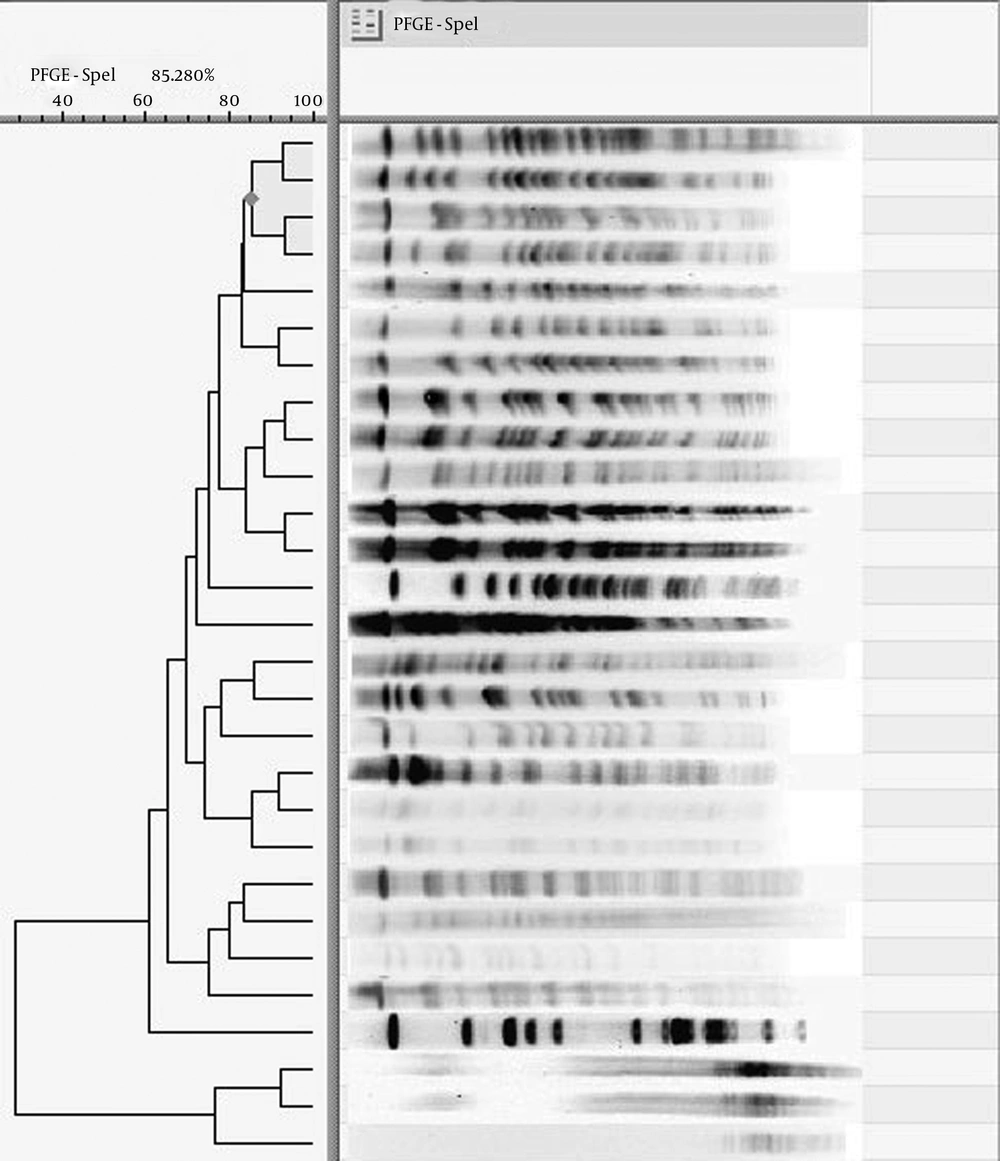
The clonal relationship among the MBL producers revealed 12 patterns: A (n = 7); B (n = 5); C (n = 1); D (n = 1); E (n = 2); F (n = 1); G (n = 3); H (n = 3); I (n = 1); J (n = 1); K (n = 2); and L (n = 1) (Figure 2). Among them, one strain was untypeable by PFGE and 6 strains had unique patterns. The pattern having the highest number of strains was defined as the dominant clone. The strains of this clone were from a variety of specimens obtained from the two hospitals in this study.
5. Discussion
A major challenge concerning P. aeruginosa infections is the ability of this bacterium to acquire resistance against new antibiotics (24). Our results showed that the isolates collected for the purposes of the present study were resistant against a variety of antibiotics. It seems that the inappropriate prescription of antibiotics in our area is one of the reasons for the increasing antibiotic resistance of this opportunistic pathogen. Furthermore, thanks to its genetic nature, P. aeruginosa is capable of acquiring a wide range of genes by means of plasmids and transposons, which allow this bacterium to become rapidly resistant against the different classes of antibiotics. The production of MBLs is one of the main mechanisms of resistance against antibiotics among P. aeruginosa isolates. Indeed, the emergence of MBL-producing P. aeruginosa isolates in hospital environments is a serious problem for the management of nosocomial infections (21). As MBL genes are located on transferable elements such as plasmids and integrons, they can easily disseminate between the strains of the bacterium.
According to recent studies in Japan, Italy, and Columbia, even without using antibiotics, P. aeruginosa isolates were capable of acquiring the VIM and IMP genes (25-27). The strains that carried these genes were also resistant to other classes of antibiotics such as Quinolones, Sulfonamides, and Aminoglycosides (28). According to epidemiological studies, the prevalence of the various patterns of drug resistance, especially the genes encoding the MBLs, can be different among P. aeruginosa isolates from one region to another or even between different hospitals in the same area (13). In Asian countries, the rates of MBL production by P. aeruginosa have been reported as 6.2% in Korea, 26.6% in Japan, and 47.3% in Taiwan (12, 29, 30). Also, studies in several parts of the world have demonstrated various rates of MBL production by P. aeruginosa, including 38.3% in Brazil, 35% in Canada, and 62% in Greece (21, 31, 32).
In our study, almost half of the isolates produced MBL enzymes, which is consistent with the above-mentioned results. Some studies in Iran have reported higher rates of MBL production among P. aeruginosa isolates (e.g.55.8% in Isfahan and 72% in Tehran) (20, 33). These differences can be explained by the geographic areas, prescribed antibiotic patterns, and the type and number of the samples tested. Among several genes encoding the MBLs, the VIM gene in P. aeruginosa is the most common (15). However, previous research has demonstrated that the frequency of this gene among P. aeruginosa isolates in Iran is low in comparison to other countries such as South Korea, Japan, and Canada (15, 33-35). In our study, the prevalence of the VIM gene was 8.3%, which is similar to the results of the previous research in Iran. Therefore, in our area, other resistant genes or mechanisms may be involved such as class D Carbapenemases, efflux pumps, and defects and loss of outer-membrane proteins including OprD (36).
The main sources of the acquisition of P. aeruginosa infections among patients have been investigated in different parts of world. Some studies have suggested that P. aeruginosa spreads from patient to patient (37), while other studies have indicated that this pathogen is predominantly acquired from the environment (38). In our study, the majority of the P. aeruginosa isolates in dominant clone A, with the highest number of strains, were from the lung specimens in both hospitals. There were strains from burn, urine, and catheter specimens in this clone as well. Given the high similarity between the genomic patterns of these strains in the current study, it can be concluded that the strains may have disseminated among the wards of each hospital and even between the two hospitals. In clone E, there were 2 strains, one from hospital A and the other from hospital B, indicating that these strains originated from a common source. Furthermore, there were 2 strains of P. aeruginosa with unique genomic patterns susceptible to Gentamicin, Piperacillin, Ceftazidime, and Cefepime which were isolated from the urine and eye specimens: they may have originated from environmental sources. The strains of clones B, C, D, E, H, I, and L were resistant to the third and fourth generations of Cephalosporins; they may have originated from a common source. The other strains, which had unique genomic patterns or contained the VIM gene, may have disseminated amongst the patients in the two hospitals from different sources.
In conclusions, our findings showed that the P. aeruginosa isolates tested were from deferent clonal origins. While some of the P. aeruginosa strains may have originated from environmental sources, the others may have disseminated among the patients in the two hospitals under study by various ways of transmission. The inter- and intra-hospital dissemination of resistant clones is a great concern and is an indicator of the level of the improvement and surveillance of standard hygiene procedures, not least disinfection and hand washing before and after contact with patients. According to our results, the resistance rate of P. aeruginosa is high and preventive strategies such as more precise antibiotic selection for treatment and less physical contact between patients should be adopted. Given the clinical significance of MBL-producing isolates, the identification of these organisms is critical if hospitals are to offer a more optimal therapeutic response and control of bacterial dissemination.