1. Background
Pseudomonas aeruginosais a frequent nosocomial pathogen that causes severe diseases in many settings, particularly in immune-compromised patients such as those with cancer, burns, and cystic fibrosis. Infections are frequently severe, and two recent studies have indicated that the rate of mortality attributable to P. aeruginosa bacteremia is approximately 34% (1, 2). This organism is clinically important since it possesses several virulence factors and is intrinsically resistant to many antimicrobial and disinfectant agents (3). Carbapenems such as meropenem and imipenem are effective antibiotics against this organism since they present a good spectrum of activity and are stable to hydrolysis by most β-lactamases, including the extended spectrum β-lactamases (3, 4). However, the use of carbapenems has been hampered by the emergence of strains resistant to carbapenems via different mechanisms such as impermeability to the drug due to loss of OprD porin, upregulation of the active efflux pump systems, and production of metallo-β-lactamases (MBLs), which hydrolyze all carbapenems (5-7).
MBLs are divided into two categories: chromosomally mediated enzymes and those encoded by movable genetic elements such as plasmids and transposons (8-10). Several kinds of MBLs have been reported, including IMP, VIM, SPM, GIM, and SIM-1 (11). The VIM and IMP types are the most clinically significant carbapenemases coded by blaVIM and blaIMP genes. At least 14 different VIM and 23 different IMP MBLs have been identified so far (7, 8). There are many reports which indicate an increase in the prevalence of carbapenem-resistance mediated by acquired MBLs; however, the prevalence of the MBL-producing isolates of P. aeruginosa varies in Asian countries (8, 12, 13). In Iran, the first isolation of blaIMP-1 was reported in 2011 by Peymani et al. (14), who collected Acinetobacter baumannii isolates from several hospitals in Tabriz (Northwest of Iran).
The growing rate of resistance against the traditional antibiotics has rendered the treatment of patients with infections caused by MBL-producing P. aeruginosa critical. Previous studies have shown a high prevalence rate of multidrug-resistant P. aeruginosa isolated from different clinical specimens, especially in burn wards in Iran (15, 16); nevertheless, little information is available on the distribution of MBL-producing isolates in the country. For example, in Markazi province, 37% of the P. aeruginosa isolates obtained from the hospitalized patients were shown to be resistant to imipenem, and 50% of these strains were detected to have blaVIM-1, 56.6% blaVIM-2, and 6.6% blaIMP-1 (17). In another study, from 240 P. aeruginosa isolates, 34.16% were imipenem-resistant, and 23.3% were screened as MBL-positive strains using the double-disk synergy test. Additionally, a specific polymerase chain reaction (PCR) test confirmed the presence of 8 (9.75%) and 10 (12.19%) IMP-1 and VIM-2 types, respectively (18).
2. Objectives
The current study was performed to examine the prevalence of MBL-producing P. aeruginosa isolates obtained from clinical specimens at different hospitals in Kermanshah (Imam Khomeini, Imam Reza, and Taleghani hospitals) and also the prevalence of those harboring the blaVIM-2 and blaIMP-1 genes, which are the most clinically significant carbapenemases.
3. Patients and Methods
3.1. Samples
From 22nd June to 22nd September 2012, 225 isolates of P. aeruginosa were collected from three hospitals in Kermanshah. The isolates were taken from urine (n = 79, 35%), blood (n = 14, 6.2%), wound (n = 29, 12.8%), respiratory tract (n = 81, 36%), burned tissue (n = 17, 7.5%), and eye (n = 5, 2.2%). The isolates were collected from different wards, including ICU, CCU, urology, emergency, burn, respiratory, and surgery. P. aeruginosa was identified by Gram staining, ability to produce oxidase and catalase, oxidative-fermentative test, resistance to cetrimide, and growth at 42°C (19).
3.2. Antimicrobial Susceptibility Testing
The disk diffusion test was conducted in accordance with the clinical and laboratory standards institute (CLSI) methodology. The antimicrobial agents included comprised aztreonam (10 µg), cefepime (10 µg), ceftazidime (30 µg), ciprofloxacin (5 µg), gentamicin (10 µg), imipenem (10 µg), and piperacillin/tazobactam (10 µg, 100 µg) (MAST, U.K.). P. aeruginosa ATCC 27853 was used as quality control in each run of antimicrobial susceptibility testing.
3.3. Identification of Metallo-β-Lactamases
Among β-lactamases, MBLs are unique in requiring the presence of zinc ion in the active site of the enzyme, and are, thus, inhibited by chelating agents such as ethylenediaminetetraacetic acid (EDTA) (20). The imipenem-EDTA double-disk synergy test was used in order to identify the P. aeruginosa producing MBLs. Therefore, a 0.50-M EDTA (Merck, Germany) solution was prepared by dissolving 186.1 g of EDTA in 1000 mL of distilled water, and the pH was adjusted to 8 by adding NaOH (21). Then, 4 μL of the prepared solution was added to the imipenem disk and dried in an incubator. The prepared EDTA/imipenem disk and the imipenem disk itself were placed in a plate containing the Müller-Hinton agar (Merck, Germany) with cultured P. aeruginosa. After 16 - 18 hours of incubation at 37°C, an increase of ≥ 7 mm in the inhibition zone diameter in the presence of EDTA compared to imipenem tested alone was considered to be a positive test for the presence of an MBL (4, 18, 22).
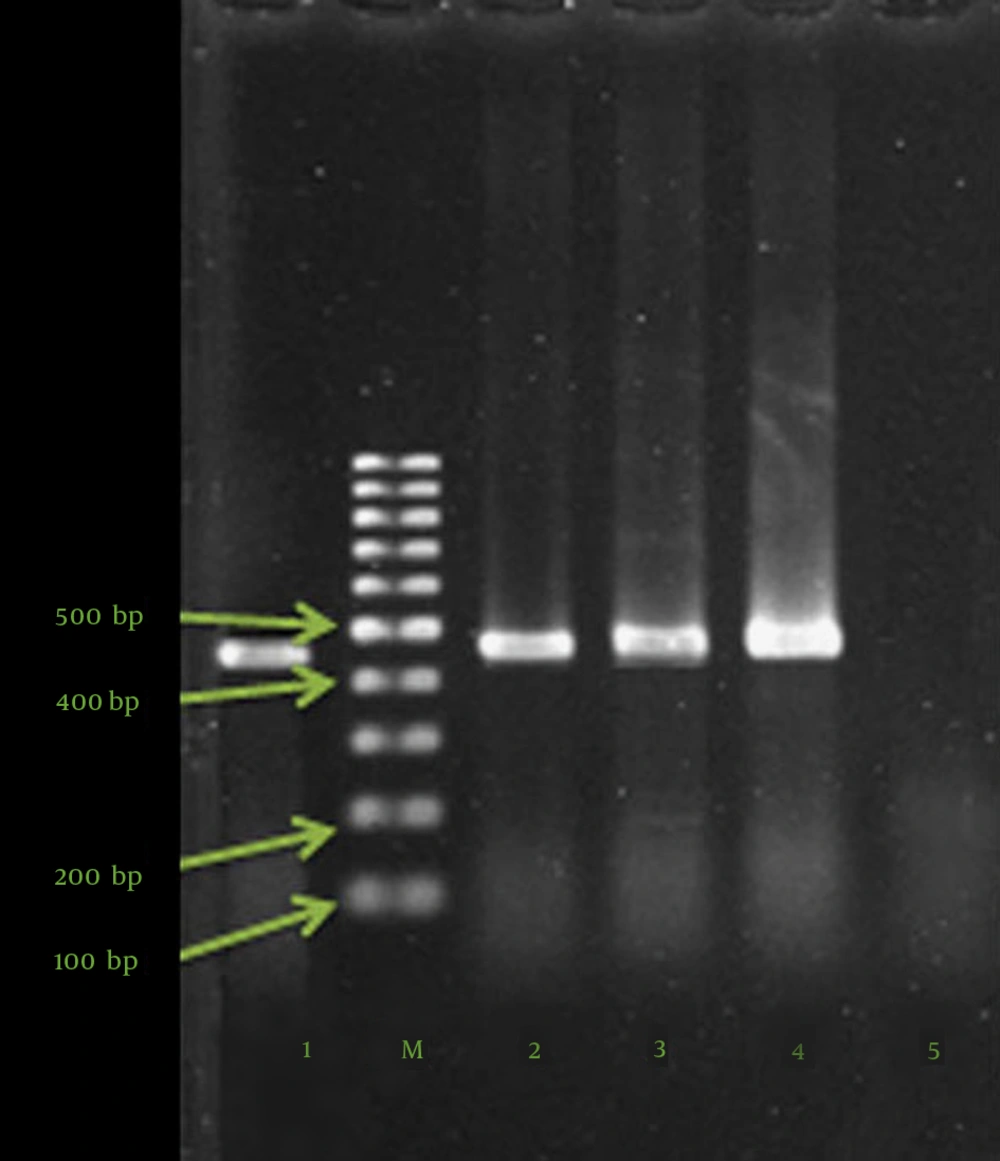
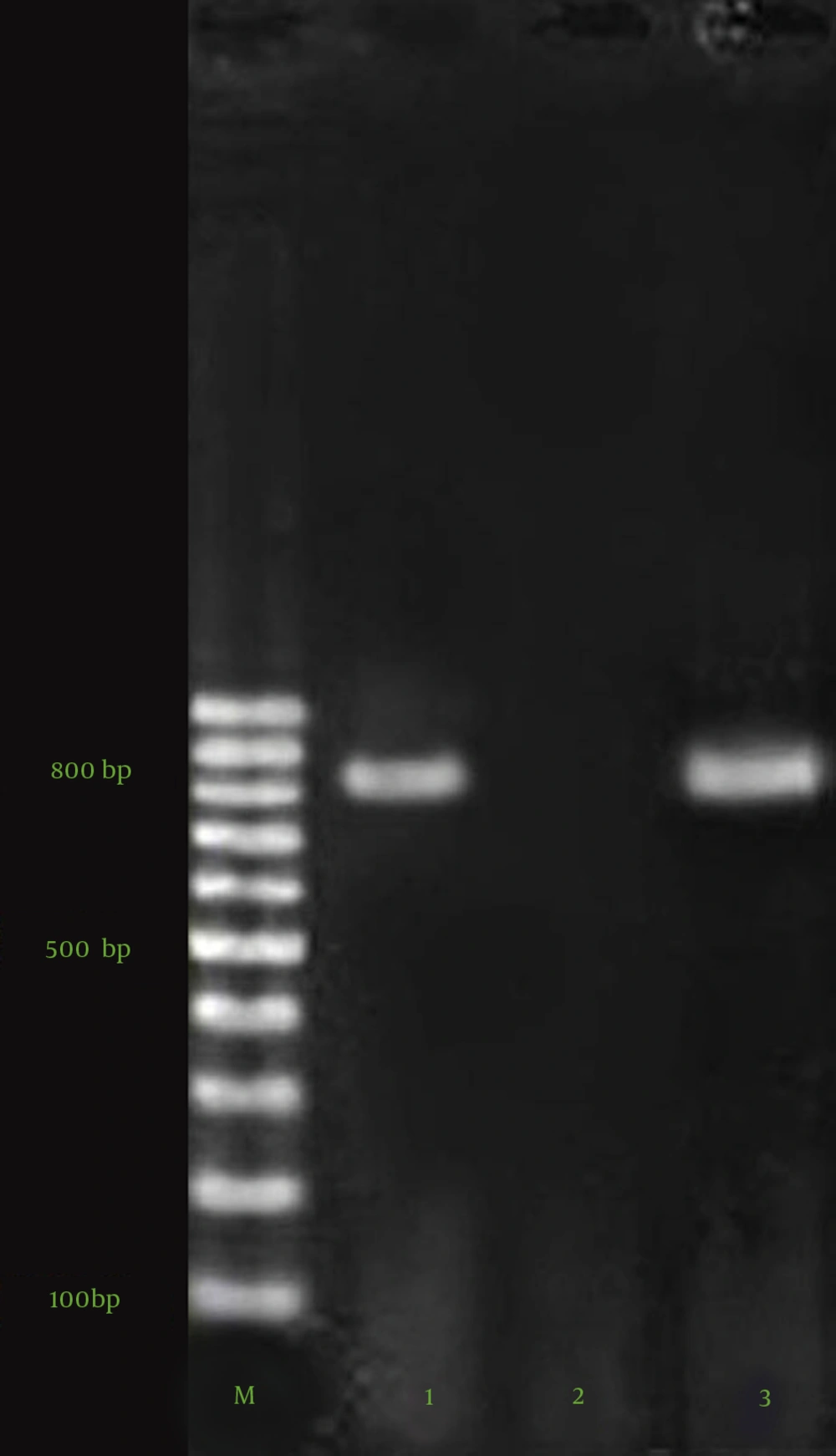
PCR was performed to detect the blaVIM-2 and blaIMP-1 genes. The DNA templates from all the isolates were extracted by boiling (10 minutes in 95°C), and PCR was carried out in a thermal cycler (Bio Rad, USA) using 25-µL volumes containing 12.5 µL of PCR Master Mix (500 mM of Tris-HCl pH 8.55, 1.5 mM of MgCl2, 0.2 mM of dNTPs, and 0.04 units/uL of Taq DNA polymerase) (SinaClon Inc., Tehran, Iran) and 0.5 µM of each of the primers (SinaClon Inc., Iran). The primers and the cycling condition are listed in Table 1 (23, 24).
Primers and Cycling Condition
Following amplification, 10 µL of each sample was analyzed with 1.5% agarose gel electrophoresis for the detection of positive samples. For all the amplification reactions, the mixture was heated at 96°C for 4 minutes prior to thermocycling. The mixture was held at 72°C for 7 minutes after the final cycle before cooling at 4°C. The agarose gel electrophoresis of blaVIM-2 and blaIMP-1 PCR products is depicted in Figure 1. The positive controls (clinical P. aeruginosa isolates with sequenced blaIMP-1 and blaVIM-2 genes) were kindly dedicated by Dr. Najjar Pirayeh (Tarbiat Modarres University), and the negative control contained water in place of DNA.
4. Results
Totally, 225 confirmed isolates of P. aeruginosa recovered from different clinical specimens were tested for antibiogram. The findings showed that 137 (61%) isolates were resistant to ceftazidime, 131 (58.5%) to gentamicin, 130 (57.7%) to piperacillin/tazobactam, 101 (44.7%) to ciprofloxacin, 96 (42.8%) to cefepime, 92 (40.9%) to aztreonam, 76 (33.7%) to imipenem, and 41 (18.1%) to meropenem. According to these percentages, imipenem and meropenem were the most effective drugs. The imipenem-EDTA double-disk synergy test showed that 45 out of the 76 (59.2%) imipenem-resistant strains were MBL positive. The isolates were collected from different wards, including ICU (n = 22), urology (n = 2), emergency (n = 4), burn (n = 11), respiratory (n = 3), and surgery (n = 3).
Furthermore, our molecular studies showed that only the MBL-positive strains were positive in PCR. Moreover, 75% (n = 34) of the MBL-positive strains were detected to have blaIMP-1 and 2.2% blaVIM-2 (n = 1). blaIMP-1 was taken from burned tissue (n = 10, 29.4 %), respiratory tract (n = 10, 29.4 %), urine (n = 8, 23.5%), blood (n = 2, 5.8%), eye (n = 3, 8.8%), and vagina (n = 1, 2.9%). blaVIM-2 (n = 1) was recovered from burned tissue. Antibiogram showed that the strains resistant to imipenem possessed high-level resistance to gentamicin (100%), cefepime (94.7%), and ceftazidime (94.7%). Among the imipenem-resistant strains, 30.3% (n = 23) were resistant to meropenem and the rest (n = 53) were sensitive to it. On the other hand, there were 18 meropenem-resistant strains sensitive to both imipenem and cefepime.
5. Discussion
Microbial resistance has increased drastically in recent years in both developed and developing countries and it has rapidly become a leading public health concern. The prevalence of antimicrobial resistance varies greatly between and within countries and between different pathogens. Resistance to antibiotics among organisms is increased either by mutations or by acquiring new genetic material via horizontal gene transfer. Several factors favor the development of bacterial resistance to antibiotics in developing countries such as the less potent activity of the drugs, i.e. some of the antibiotics provided in developing countries have decreased potency due to the degradation or adulteration of the drug or because of the presence of a lower concentration of active substances, lack of well-equipped diagnostic laboratories, and excessive use of antimicrobials in domestic animals (8-10).
Antimicrobial agents suitable for use in pediatric clinics are mainly limited to β-lactam antibiotics, especially carbapenems. However, the problem of drug resistance to carbapenems is gradually worsening owing to their increasing clinical use. The present study analyzed the patterns of resistance to several antibiotics such as carbapenems. The results showed that 33.7% of the isolates were resistant to imipenem. In a previous study in the Iranian city of Kermanshah in 2004, Mohajeri et al. (25) reported a lower rate of resistance (10%). However, another investigation conducted in the Iranian city of Shiraz in 2012 reported that 127 (59.16%) of the 240 isolates were resistant to this antibiotic (18). The reported rates of resistance to imipenem in Japan, Russia, France, Canada, and Spain were 8.3%, 13.4%, 18.5%, 12%, and 14%, respectively (26-30). As was mentioned, numerous factors contribute to these variations. In addition, the increased rate of resistance to imipenem in Kermanshah is in line with the increase throughout the world. Resistance to carbapenems in P. aeruginosa may be due to MBLs, which hydrolyze all carbapenems (5-7). Since MBLs are placed in movable elements, they are horizontally transmitted between organisms easily (8-10, 31). Large outbreaks by MBL-producing P. aeruginosa strains have been described in hospitals in Italy, Greece, and Korea (32-37). IMP and VIM-producing P. aeruginosa strains have been reported worldwide (10, 38-42).
In our study, the majority of the MBL producers carried the blaIMP gene (75%); however, the presence of this gene has been previously proved to be variable in different regions (17, 18). In a previous report from the Iranian province of Markazi, 37% (40 out of 108) of the P. aeruginosa isolates obtained from the hospitalized patients were demonstrated to be resistant to imipenem. The EDTA/imipenem test showed that 50% of the imipenem-resistant strains were MBL positive, and PCR revealed that 50% of the imipenem-resistant strains had blaVIM-1, 56.6% blaVIM-2, and 6.6% blaIMP-1 (17). In another study in Iran, from 240 P. aeruginosa isolates mainly recovered from wound, urine, and sputum, 82 (34.16%) isolates were imipenem-resistant (minimum inhibitory concentration [MIC] ≥ 4 μg/mL). Among these imipenem-resistant isolates, 19 (23.3%) MBL-producing P. aeruginosa isolates were screened using the double-disk synergy test. A specific PCR test confirmed the presence of 8 (9.75%) and 10 (12.19%) IMP-1 and VIM-2 types, respectively (18). In contrast, Khosravi and Mihani (2008) reported that none of the 8 MBL-producing P. aeruginosa strains in their study were positive for the blaIMP gene (40).
The most and also the sole frequent reported MBL-producing P. aeruginosa in Iran is VIM-1, previously cited by several researchers in different cities of Iran (4, 17, 22, 40, 43, 44). Nonetheless, there are only a few investigations reporting the presence of the VIM-2 type of P. aeruginosa in Iran. For the first time in 2012, Sadeghi et al. (17) detected this genotype in the Iranian city of Arak. Subsequently, in 2013, among the 82 imipenem-resistant isolates tested for MBLs, 10 isolates were positive for this gene (18). According to our findings, it seems that the MBL-producing isolates of P. aeruginosa are the main cause of IPM resistance among this species. We suggest that molecular typing via standard methods such as pulsed-field gel electrophoresis be conducted to define the clonality of the isolates.