1. Background
Leptospirosis occurs both in developing and industrialized countries. Wild and domestic animals are important carriers of the disease (1). Today, leptospirosis is recognized as a re-emerging infectious disease; therefore, understanding its epidemiology is a vital issue for designing intervention programs and diminishing its transmission. The disease has peak incidence during rainy seasons in warm climate regions and in summer or fall in temperate regions (2, 3). The first leptospirosis outbreak was reported in Iran by Tahbaz (4). Examination of leptospirosis in livestock around Tehran in 1992 revealed dominant serovars of Leptospira interrogans as Hardjo, Tarasovi, Grippotyphosa, Pomona, and Icterohaemorrhagia. On the other hand, the most serovars isolated from humans and cattle throughout Iran in the last 5 years were Grippotyphosa, Canicola, Hardjo, and Icterohaemorrhagia that were identified by Razi Institute Researchers (5).
Identification of Leptospira species has been conventionally performed by cultural and serological methods. These methods are tedious, time consuming, and potentially biohazardous. For example in microagglutination test, live cultures of strains must be kept for use for their antigens, therefore laboratory facilities are required. Besides, the results are difficult to standardize and need a biologist to operate the microscope. Above all, cross-reactions occur between serogroups, that makes the interpretation of the results complicated (6). Recently, other molecular methods such as pulse field gel electrophoresis (PFGE) and arbitrarily primed PCR (AP-PCR), restriction fragment length polymorphism (RFLP), multilocus variable number tandem repeat analysis (MLVA) are introduced (7-11). MLVA method provides useful tools in detection and identification of Leptospira serovars and gives information relating to both the evolutionary and functional areas of bacterial diversity. Leptospiral genomes contain multiple loci of repetitive DNA, which may be variable among serovars with respect to their individual primary structure or number of repeat units present. The variation in the number of repeats arise from slipped strand mispairing as proposed for other short sequence repeats (12).
2. Objectives
The aim of this study was genomic identification of pathogenic Leptospires in Iran by multilocus variable-number tandem-repeat analysis.
3. Materials and Methods
3.1. Bacterial Serovars and Culture Conditions
Leptospira interrogans serovars (Table 1) were obtained from Leptospira Reference laboratory of Microbiology Department, Razi Vaccine and Serum Research Institute, Karaj, Iran. These serovars were isolated from humans and cattle during last year and maintained in the institute. Leptospira serovars were grown in EMJH liquid medium (Difco) at 28˚C for up to 3 weeks to reach the stationary phase (13, 14).
| RTCC No. | Species | Serogroup | Serovar | Strain | Abbreviation |
|---|---|---|---|---|---|
| 2802 | L. inerrogans | Autumnalis | Autumnalis | Akiyami A | Aut |
| 2805 | L. inerrogans | Canicola | Canicola | Hond Utrecht IV | Ch1 |
| 2808 | L. inerrogans | Grippotyphosa | Grippotyphosa | Moskava | G1 |
| 2810 | L. interrogans | Sejroe | Hardjo | Hardjo bovis | Sh1 |
| 2812 | L. inerrogans | Icterohaemorrhagia | Icterohaemorrhagia | Verdum | Ict1 |
| 2815 | L. inerrogans | Pomona | Pomona | Pomona | Po1 |
| 2817 | L. inerrogans | Sejroe | Serjae serjae | M84 | Ser |
| 2819 | L. biflexa | Semaranga | Patoc | Patoc1 | Pat |
| 2821 | L. interrogans | Sejroe | Hardjo | Hardjo-prajitno | Sh2 |
| 2822 | L. inerrogans | Pomona | Pomona | UT364 | Po2 |
| 2823 | L. inerrogans | Icterohaemorrhagia | Icterohaemorrhagia | RGA | Ict2 |
| 2824 | L. inerrogans | Canicola | Canicola | Fiocruz LV133 | Ch2 |
| 2825 | L. inerrogans | Grippotyphosa | Grippotyphosa | Andaman | G2 |
a The data are presented by Razi Type Culture Collection.
3.2. VNTR Primer Selection
MLVA serovar typing was performed with the VNTR4 primer designed and used by Salaun et al. (14); VNTR7 and VNTR10 described by Majed et al. (13); VNTR8 and VNTR29 defined by Slack et al. (15) (Table 2).
| VNTR Locus | Sequences 5’→3’ | Reference |
|---|---|---|
| VNTR4 | 14 | |
| Forward | (AAGTAAAAGCGCTCCCAAGA) | |
| Reverse | (ATAAAGGAAGCTCGGCGTTT) | |
| VNTR 7 | 13 | |
| Forward | (TCATCTGCTCCGGAGATTCG) | |
| Reverse | (TCCCTCCACAGGTTGTCTTG) | |
| VNTR10 | 13 | |
| Forward | (TCCAAAATTCAGCCCTCAAG) | |
| Reverse | (GACGCTTGGCATTTGTATCC) | |
| VNTR8 | 15 | |
| Forward | (CAAGTGTTCGACAAGAATGAG) | |
| Reverse | (CTCACCGGTAGAACGCTTCTTTT) | |
| VNTR29 | 15 | |
| Forward | (ATCGTTTTGGCAGTTTTTGCT) | |
| Reverse | (CTAGAAAATTCCGCGTAGGG) |
3.3. DNA Extraction and PCR
The genomic DNA of Leptospira was extracted by phenol-chloroform method (Merck, Germany). The final volume (25 µL) of each reaction mixture contained 2.5 µL of 1X PCR buffer (SinaClone, Iran), 0.5 mM of 10 mM deoxynucleoside triphosphate (SinaClone, Iran), 1 µM of 10 pmol each corresponding primer, 0.75 of 50 mM MgCl2 (SinaClone, Iran), 0.3 unit/µL of 0.5 unit Taq DNA polymerase (SinaClone, Iran) and 1 µL of 100 macro gram DNA template. Amplification was achieved under the following conditions: 1 denaturation cycle at 93˚C for 5 minutes; 35 cycles of denaturation at 93˚C for 1 minute, annealing at 54˚C for 1 minute, elongation at 72˚C for 1 minute, and a final elongation at 72˚C for 10 minutes (14). The amplified products were analyzed by 2% agarose gel electrophoresis and allelic sizes estimated by Quantity One 1-D analysis software package (Bio-Rad, USA). The size of the amplified products was estimated by comparison with a 100 bp plus ladder (15, 16). Some of the amplified products were sequenced by Macrogene Company in South Korea.
3.4. Sequence Analysis
The copy number of repeats of each VNTR locus was deduced from sequencing data and sizes of the amplified products. From 12 strains and 3 VNTRs, 36 fragments were sequenced. The ClustalX program was used to generate nucleotide sequence alignments. Then, allelic sizes were converted in to repeat copy numbers using Microsoft Excel software package by the following formula:

Next, the repeat copy numbers were rounded down to form whole numbers (Table 3). Clustering analysis was done using the categorical parameter and the ward coefficient. Nei’s diversity index of the VNTR loci was calculated from the range of alleles generated from the reference strains utilizing the formula (Table 3) (14-17):

| VNTR locus | Repeat Motif | Repeat Size, bp | Total Flanking Region | Repeat Range, Min - Max | Total Length of PCR product | Nei’s Diversity Index |
|---|---|---|---|---|---|---|
| VNTR4 | TCGAGCGCCCATAGAAGCGAGACGCTGAGTTACT | 34 | 352 | 1 - 4 | 428 - 526 | 0.69 |
| VNTR10 | CAGTTAAATTTAGAAATGTGGGAACACACACCATTTGCGGGTTTTA | 46 | 72 | 4 - 13 | 296 - 710 | 0.65 |
| VNTR29 | GATTTTACAGTTAGACTTTGAAATTGTGGGAACTCCCACGGATTTGG | 47 | 28 | 3 - 10 | 209 - 538 | 0.6 |
4. Results
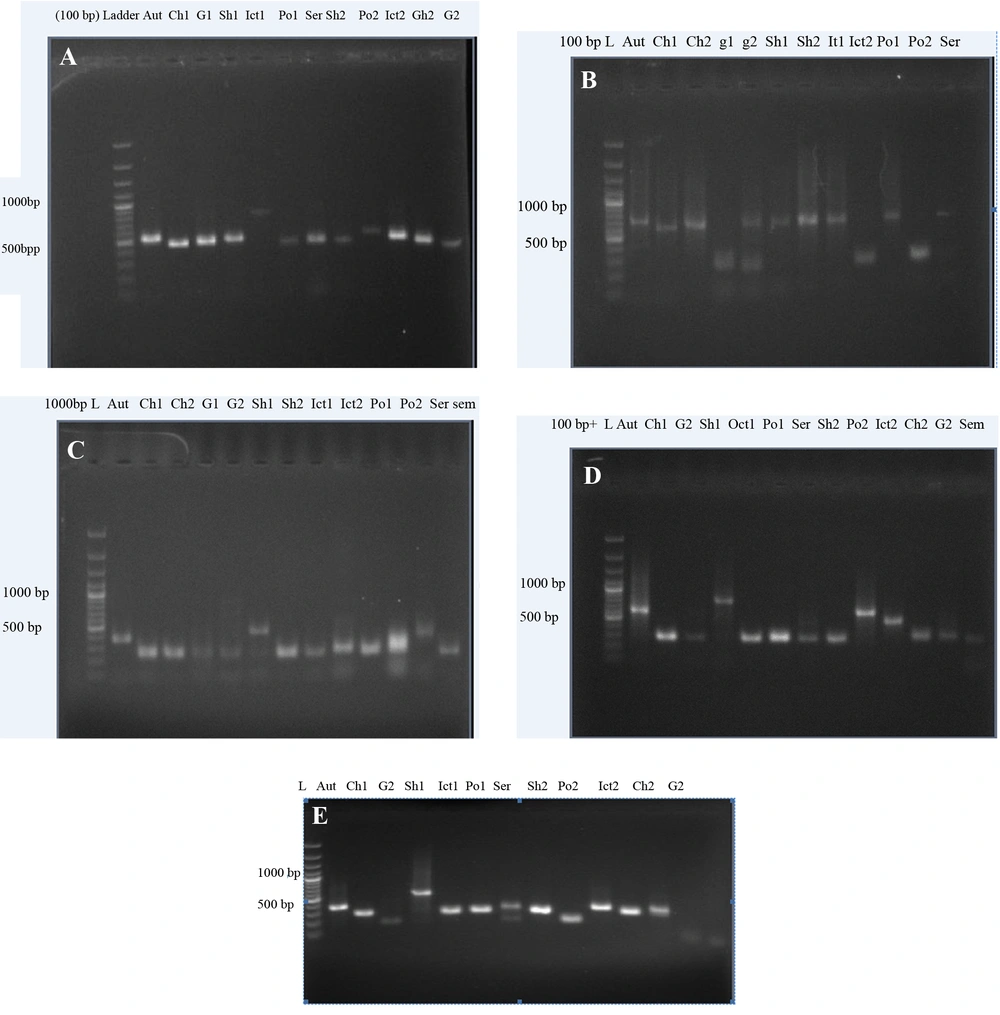
Analysis of the amplified PCR products revealed size variations in most of the loci by agarose gel electrophoresis. All loci exhibited a single PCR product whose size could be determined by 2% agarose gel electrophoresis. All loci successfully amplified in all pathogenic Leptospira serovars. The saprophytic serovar showed no amplified fragments. The sizes of the amplified products displayed range of polymorphism, suggesting variation in tandem repeat copy numbers in 5 VNTR loci (Figure 1). This was confirmed by sequencing of 36 amplified products from three out of five loci (VNTR4, VNTR10, and VNTR29 out of all).
Amplification was performed on the VNTR4 (A), VNTR7 (B), VNTR8 (C), VNTR10 (D), and VNTR29 (E) (full names of the abbreviation of the serovars are listed in Table 1). For each VNTR locus, sequence analysis of amplified products displayed a high conservation of flanking regions and repeat units among L. interrogans serovars (Table 3). For each locus, the number of tandem repeats was calculated by measuring the sizes of the amplified products. The strains were typed by the numbers of variable tandem repeats in each of 5 VNTR loci. Among VNTR sequences analyses, VNTR 4 was thoroughly differentiated between L. interrogans serovar (Grippotyphosa strain Moskava, Hardjo strain Hardjo bovis, Pomona strain UT364, and Icterohaemorrhagia strain RGA) from other serovars. In fact, VNTR 4 detected 6 alleles through 13 serovars. VNTR 10 and VNTR29 had an appropriate discrimination to differentiate 5 alleles among 13 serovars (Table 4).
| Serovars | RTCC No. | VNTR4 | VNTR10 | VNTR29 |
|---|---|---|---|---|
| Autumnalis | 2802 | 526 | 618 | 350 |
| Canicola | 2805 | 460 | 342 | 303 |
| Grippotyphosa | 2808 | 479 | 296 | 209 |
| Hardjo | 2810 | 495 | 710 | 538 |
| Icterohaemorrhagia | 2812 | 460 | 342 | 303 |
| Pomona | 2815 | 460 | 342 | 303 |
| Serjae serjae (2817) | 460 | 342 | 317 | |
| Patoc | 2819 | - | - | - |
| Hardjo | 2821 | 460 | 296 | 303 |
| Pomona | 2822 | 562 | 618 | 209 |
| Icterohaemorrhagia | 2823 | 526 | 526 | 350 |
| Canicola | 2824 | 460 | 342 | 303 |
| Grippotyphosa | 2825 | 428 | 342 | 303 |
a The values are presented as bp.
5. Discussion
Leptospirosis has a global distribution and is more prevalent in tropical regions than in temperate zone. This is due to longer survival of leptospirosis (18, 19). Currently, leptospirosis has spread from rural regions to epidemics in urban communities. Understanding the epidemiology of this phenomenon is a vital issue for designing intervention programs and subsequent diminishing its transmission. Unfortunately at present there are few prevention measures for leptospirosis (3).
Today, MLVA method is used for differentiation and identifying Leptospira serovars. The method has potential application in understanding the leptospiral molecular epidemiology with its easy, rapid, and high discriminatory power. Besides, this method is convenient for the exchange of information between laboratories (14, 15). Compared to other bacteria, L. interrogans genome displayed a larg number of tandem repeats with a total length greater than 100 bp for strains within a species or genus (14). The saprophytic serovar was used in this study showed no amplified fragments (the same results were obtained by Majed et al. (13) Slack et al. (15, 16) Salaun et al. (14) and Pavan et al. (17) for other saprophytic serovars). So maybe they are good markers for separation between pathogenic and non-pathogenic serovars and also these loci may interfere in the pathogenesis and virulence.
The combination of VNTR4, VNTR7, VNTR10, VNTR8, and VNTR29 were useful for the differentiation of L. interrogans serovars. It seems that VNTR method can provide rapid typing as well as a highly discriminatory assay to identify L. interrogans serovars in large scale for epidemiological investigation. This finding was in agreement with Majed et al. (13) study that demonstrated high range of polymorphism of VNTR7 and VNTR10 among all VNTR loci. Also, it is in line with the study of Pavan et al. (17) who claimed to set the groundwork for regional Leptospira genotype databank, and provide essential information for future molecular diagnosis and epidemiological tracing of Leptospira by MLVA.
Furthermore, some serovars for each locus displayed the same patterns (Table 4). For instance, VNTR4 has the same pattern in L. interrogans serovar Canicola, strain Hond utrecht IV; serovar Icterohaemorrhagia, strain Verdum, serovar Serjae Serjae; serovar Hardjo, strain Hardjo prajitno; and serovar Canicola, strain Fiocruz LV133. So it seems that serovars from the same geographical area could have more genetic similarity than the serovars in different places such as South America and Europe. The same results were obtained by Majed et al. (13) and Slack et al. (15, 16). They also showed strains originating from distant continents can be grouped together in the dendrogram (13).
One important point regarding MLVA is its ability to show genetic diversity within a few same serovars of the collection strains underestimating the variety of isolates in natural populations, the point that PFGE was unable to differentiate. For instance, among serovars of Hardjo St; Hardjo bovis and Hardjo prajitno and among Pomona St.; UT364, Pomona St. Pomona heterogeneity were identified by MLVA, but PFGE was unable to differentiate them. It was similar to Salaun et al. (14) results. She found among the 5 L. kirschneri strains from Guadeloupe (French West Indies islands) L. kirschneri, serovar Bogvere by PFGE, and two genotypes by MLVA. Similarly, among the 4 L. kirschneri strains identified as L. kirschneri serovar Grippotyphosa by PFGE, MLVA identified different patterns. This finding suggests that a high heterogeneity exists among serovars identified as Grippotyphosa by PFGE (14).
The VNTR analysis developed in this research was cost-effective and easy to perform. Combination of VNTR4, VNTR7, VNTR10, VNTR8, and VNTR29 were useful for differentiation of L. interrogans serovars. This technique provides a base on which improvements to the method and comparisons to other methods can be made. These results highlight the potential role of MLVA in molecular epidemiology of L. interrogans serovars.