1. Background
Haemophilus influenzae is an important pathogen among infants and children. The serotype b strains of H. influenza (Hib) are a major cause of invasive infections (1). Nontypeable Haemophilus influenzae (NTHi) is a frequent commensal of the human nasopharynx but is also the common cause of respiratory tract infections such as otitis media, sinusitis, bronchitis, and pneumonia (2, 3). Although effective vaccines against the Hib strains have been used widely (4), they do not protect children against infections caused by the NTHi strains. The prevention of NTHI infections would provide considerable health and economic benefits. Thus, to develop a vaccine that protects against Hib and NTHi infections, several surface-exposed H. influenzae proteins such as pili and outer membrane proteins have been intensely studied (5-8). Vaccine candidate selection for H. influenzae is not easy because NTHi demonstrates extensive sequence and antigenic variation among the gene products interacting with the immune system such as outer-membrane proteins, adhesins, lipopolysaccharides, and secreted virulence factors (9-12). One of the possible candidates of a vaccinogen is protein D (PD) (3). The antigenic conservation of PD and the role of this protein in the onset of H. influenzae infection suggest that PD is a candidate antigen for a vaccine to prevent nonencapsulated H. influenzae infection (13). PD manifests glycerophosphodiester phosphodiesterase activity, which is required for the transfer of choline from the host to the lipooligosaccharide of H. influenza (14-16). PD has also been proven to promote bacterial adhesion and internalization into human monocytes (17).
2. Objectives
The aim of the present study was to design a new truncated form of PD, to predict its B cell epitope, and to perform a protein structure modeling of the truncated form using bioinformatic tools with a view to assessing this constructed recombinant truncated PD as a vaccine candidate against H. influenzae. We used an expression system to produce this protein in Escherichia coli on a laboratory scale with the potential of production on an industrial scale. Further studies should be performed in order to evaluate the immune system.
3. Materials and Methods
3.1. In Silico Design
The truncated PD design was based on multiple sequence alignment of full-length protein sequences from several H. influenzae in the GenBank using ClustalW Multiple Sequence Alignment software, and the conserved areas of the PD sequence of H. influenzae were also selected. We used the immune epitope data base (IEDB) analysis resource (http://www.iedb.org) to identify the immunogenic epitopes of the H. influenzae PD. The modeling of the truncated protein was determined by I-TASSER website. The result of the modeling was validated and analyzed using protein structure analysis ProSa (https://prosa.services.came.sbg.ac.at/prosa.php) and SPDBV software Z-score (overall model quality). The Ramachandran Z-score (for calculating the quality of a Ramachandran plot) was calculated by using the SPDB Viewer.
3.2. DNA Isolation
Plasmid DNA was prepared by using a Qiagen plasmid DNA kit (Diagen GmbH, Dusseldorf, Germany) according to the instructions of the manufacturer. The genomic DNA of the H. influenzae strain ATCC49766 was prepared by using a genomic DNA extraction kit. Bacterial strains were routinely grown at 37°C in lysogeny broth (LB) broth or agar (Merck, Germany), supplemented with 50 μg/mL of ampicillin as required.
3.3. Primers Design and Polymerase Chain Reaction
The truncated hpd gene was amplified from the chromosomal DNA of the H. influenzae strain ATCC49766 via Polymerase Chain Reaction (PCR). Oligonucleotide primers were prepared on the basis of the published nucleotide sequence of the hpd gene from NTHi. The primers were designed based on the truncated hpd gene of the 86-028NP strain (GenBank accession nos. CP000057.2) with NcoI and XbaI restriction sites (underlined), respectively. The sequences of the primers were as follows:
F: 5’-CAT GCC ATG GAA GAA ACG CTC AAA G-3’
R: 5’-GAT CTC TAG AGC ATT ATC AGG TTT GGA TTC TTC-3’
The PCR reactions were performed using the Eppendorf thermocycler. The PCRs were carried out in a 50 µL volume containing 2 µlL of DNA template, 5 µL of 10x reaction buffer, 2 µL of dNTPs (10 mM), 2 µL of MgCl2 (50 mM), 2 µL of each primer (10 pmol), and 1U of pfu DNA polymerase (Fermentas). Amplification was performed by Hot Start at 95˚C for 3 min, followed by 35 cycles of denaturation at 94˚C for 10 sec, annealing at 50˚C for 15 sec, extension at 72˚C for 90 sec, and 10 min at 72˚C for final extension. The PCR product was recovered from the gel and purified with a high pure PCR product purification kit (Fermentas) according to the manufacturer’s recommendations.
3.4. Cloning, Construction of the Recombinant Protein and Sequence Analysis
The PCR product was digested with NcoI and XbaI and ligated into pBAD that had been digested by the same enzymes, which provided 6 His residues at the N-terminus of the expressed protein. The ligations were performed with T4 DNA ligase. The ligation mixtures were transformed into competent E. coli TOP10 by the CaCl2 method. The transformed bacteria were cultured on LB agar containing 50 μg/mL of ampicillin, and selected colonies were analyzed to present the construct by PCR and restriction enzyme digestion. Finally, the sequencing of the constructed plasmid was performed by specific pBAD universal primers.
3.5. Expression of the Truncated Gene and Purification of the Recombinant Protein
Recombinant E. coli TOP10 cells were grown overnight in a Luria broth (LB) medium containing ampicillin (50 µg/mL) at 37°C. On the following day, 50 mL of the LB medium was inoculated with 0.5 mL of the overnight culture of TOP10. The inoculated culture was grown by agitation under aerobic conditions at 37°C to an A650 of 0.5 - 0.6. Then, the expression of the cloned gene was induced by different concentrations of Arabinose (final concentration 0.02% - 20%). After incubation for 4 hours, the cells were harvested by centrifugation at 4°C and were stored at -20°C until further use. Protein was expressed and purified by using the Ni-NTA column (Qiagen) under native conditions according to the manufacturer’s instructions. The quality and quantity of the purified recombinant PD was analyzed on a 15% SDS-PAGE gel electrophoresis by the Bradford method.
3.6. SDS-PAGE and Western Blot
The expression of the proteins was analyzed by SDS-PAGE. The bacterial pellets were suspended in a loading buffer, heated for 5 minutes at 95°C, and 25 µL of each sample was subjected to 15% SDS-PAGE gel. For the western blot, the samples were separated by the SDS-PAGE and transferred in nitrocellulose membranes by using a liquid transfer system (Bio-Rad). The membranes were blocked with skimmed milk in phosphate buffered saline (PBST) (PBS 1% + Tween 20) and then washed several times with PBST. The membranes were incubated with the conjugated His-tag antibody (Roche) for 1 hour at room temperature and developed by Diaminobenzidine (DAB) solution (Roche, Germany).
4. Results
4.1. In Silico Design
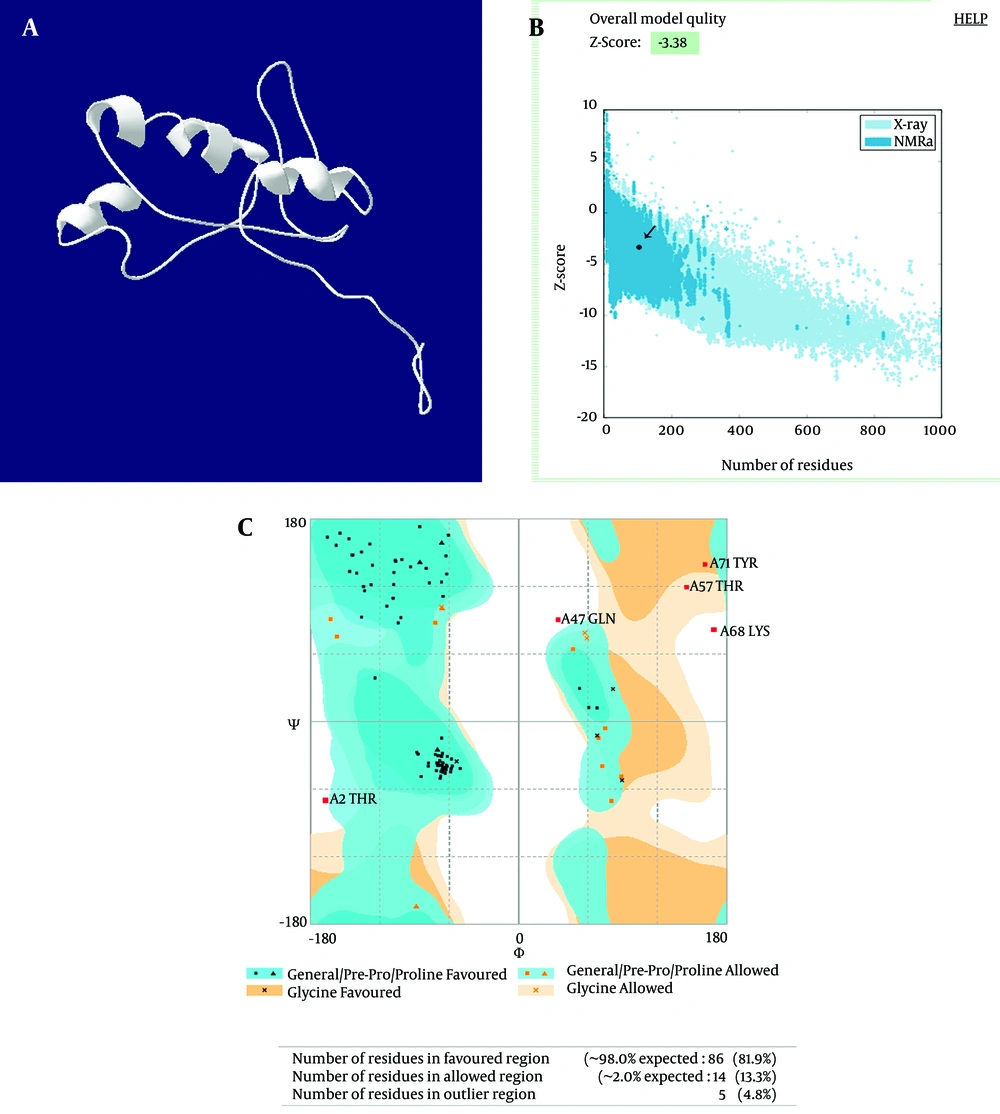
Multiple sequencing alignments of the hpd gene among the H. influenzae sequence in the GenBank showed 90% homology. The results of mouse B cell epitope prediction showed that the peptides were located in the highly conserved area. Based on the alignment and epitope prediction results, the amino acids 192 to 299 formed the truncated form. The structure Z-score and the Ramachandran Z-score of the truncated PD structure were 3.38 and 98%, respectively. The Z-score indicated that it was in the range of native conformations, and the plot showed that the structure was within the range of scores, typically found for native proteins of a similar size (Figure 1).
Showing (a) truncated protein D (aa 192-299) structure prediction by I-TASSER server. (b) The Z-score plot of the truncated protein D shows that the structure is within the range of scores typically found for native proteins. (c) The Ramachandran plot of the truncated protein D shows that the protein structure has 100% of its residues in most favored areas, resulting in Z-scores.
4.2. Amplification of the Truncated Protein D and Construction of the pBAD-Truncated Protein D
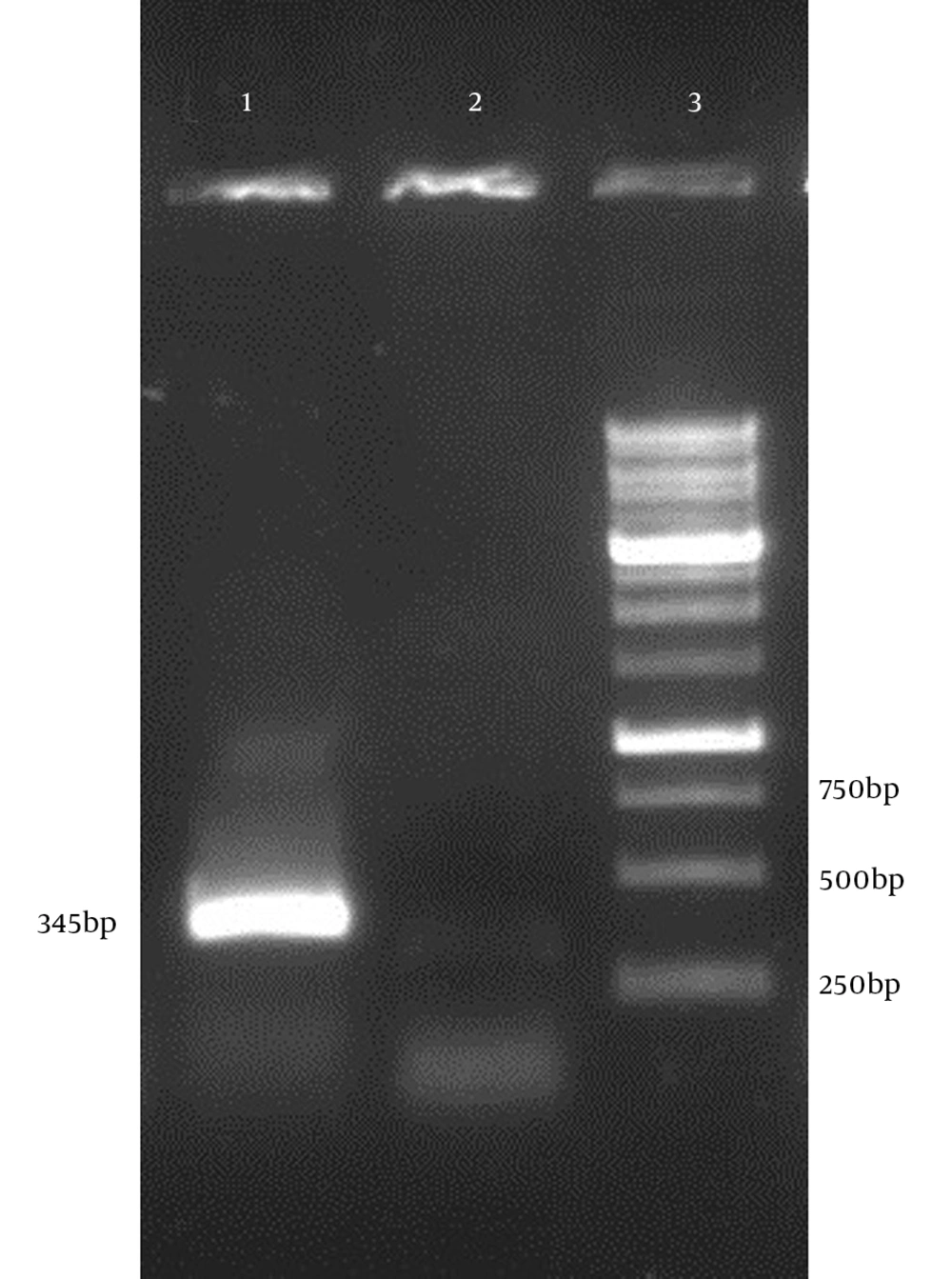
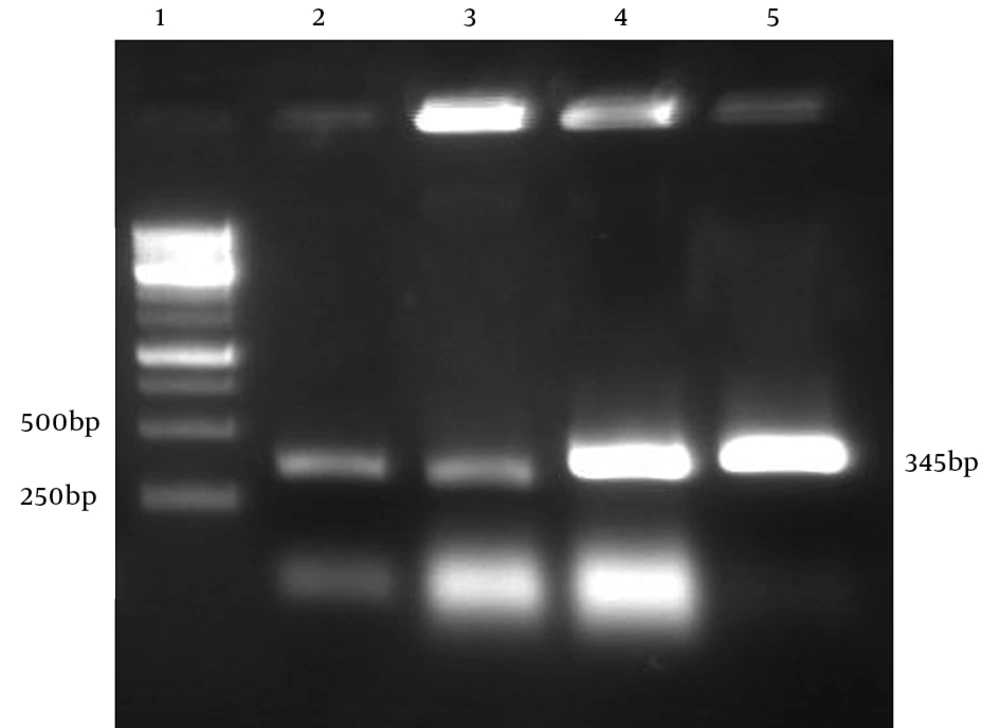
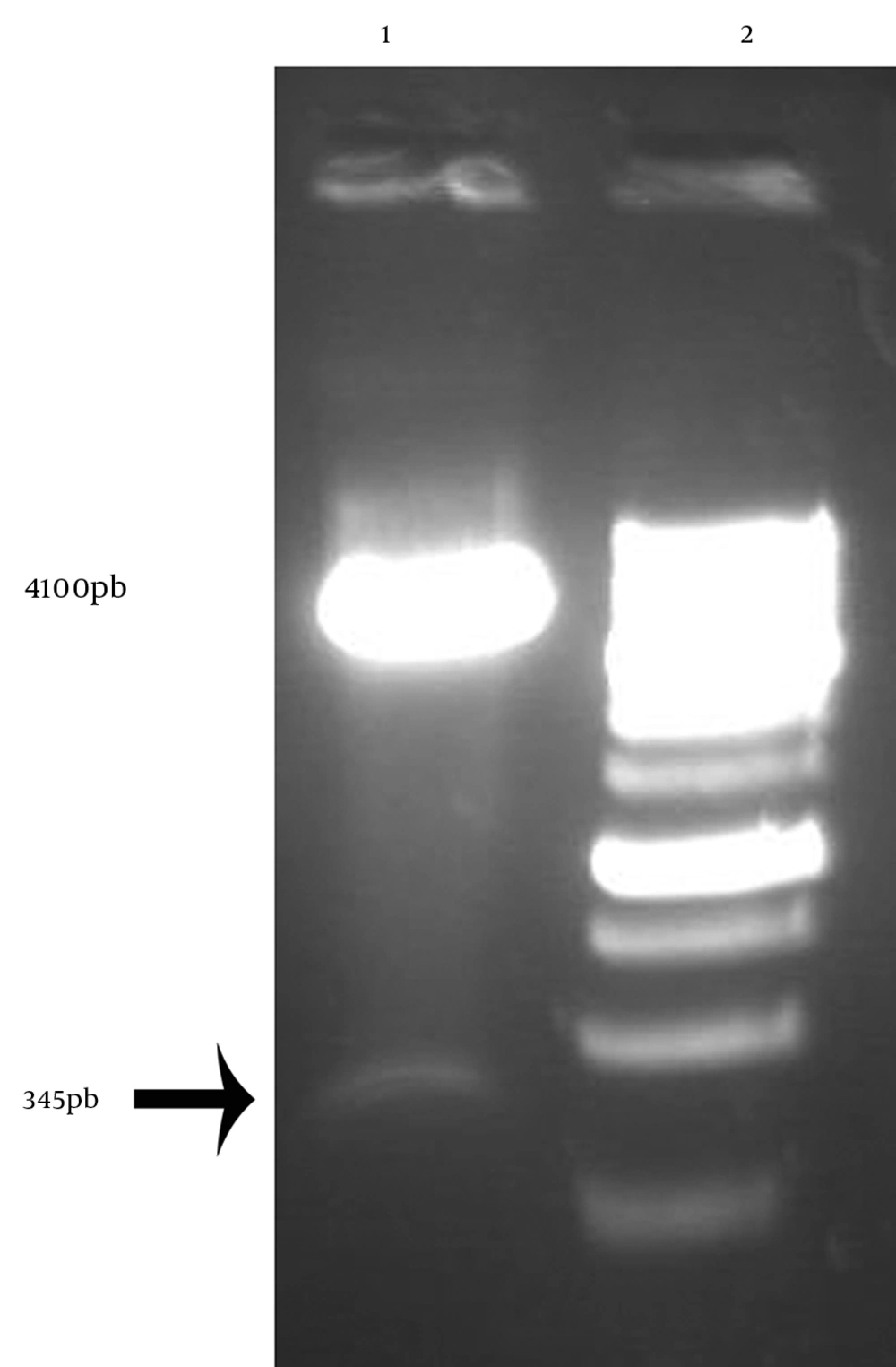
The PCR conditions were optimized for amplification of truncated hpd gene. Electrophoresis of PCR products showed that the length of PCR fragment of truncated hpd gene was approximately 345 bp (Figure 4). Double digestion DNA was ligated into digested expression vector pBAD. E. coli Top10 cells, transformed with the ligation mixture. Individual colonies were screened to find if they were positive by colony PCR(Figure3). The positive colonies were picked up and purified. The restriction enzyme analysis of the plasmid DNA from the positive clones confirmed the presence of the insert (Figure 4).
The positive colonies were picked up and purified. The restriction enzyme analysis of the plasmid DNA from the positive clones confirmed the presence of the insert (Figure 4).
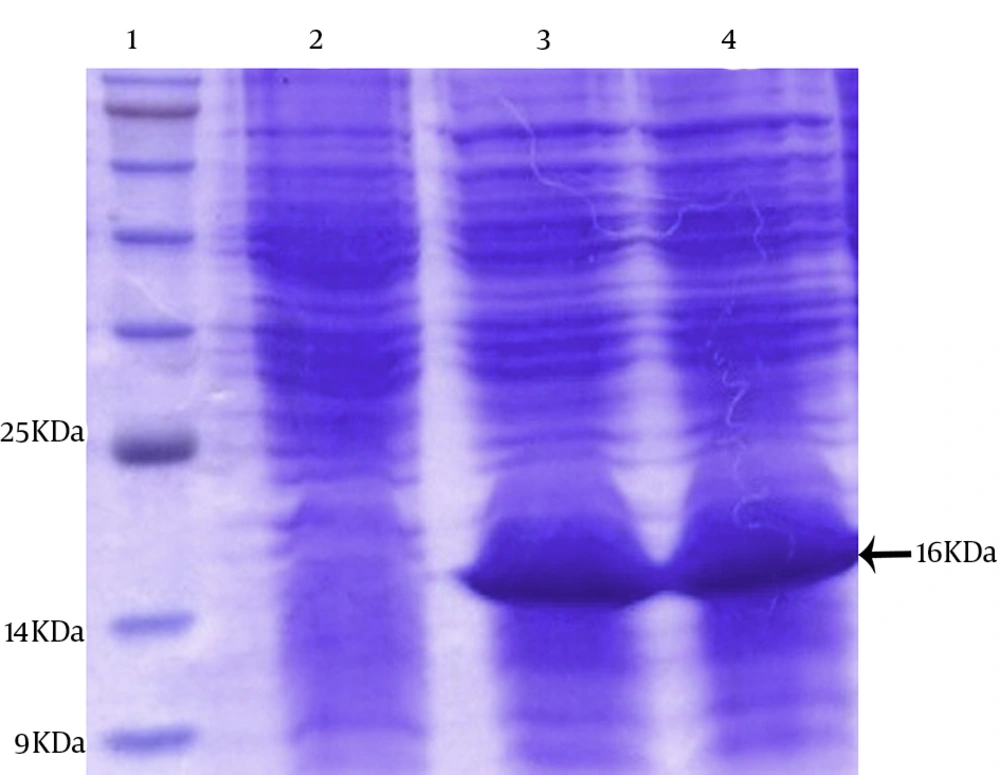
4.3. Expression of the Truncated hpd Gene Cloned and Purification of the Recombinant Protein
The pBAD was optimized by parameters such as different concentrations of Arabinose and incubation time. Optimum expression was obtained with 0.2% Arabinose and an incubation time of 4 hours. The TOP10 cells harboring constructs were cultured at 37°C in the presence and absence of an inducer Arabinose. The whole-cell lysates were analyzed by 15% SDS-PAGE. One major band appeared approximately at the 16 kDa position after Arabinose induction. The induction of the cells with Arabinose (2%) at 37°C for 4 hours was found to be optimal to achieve high-level expression (Figure 5).
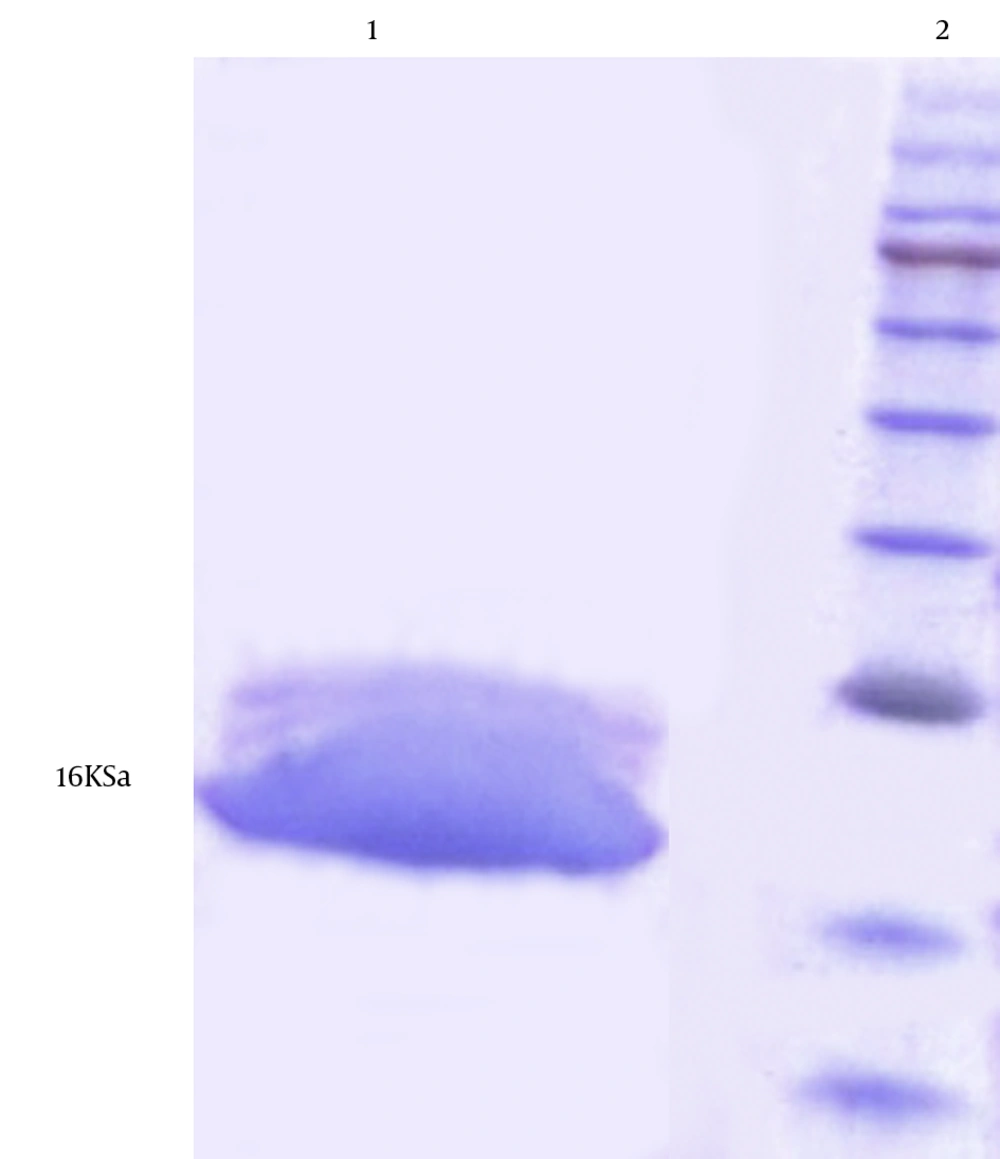
The expressed protein was purified successfully via affinity chromatography using Ni-NTA resin HYPERLINK “http://jjmicrobiol.com/?page=article&article_id=10009#figf10309” (Figure 7 ). The purification and dialysis process resulted in the yield of about 30 mg of purified protein from 1 L of E. coli Top10 + pBAD-truncated D culture.
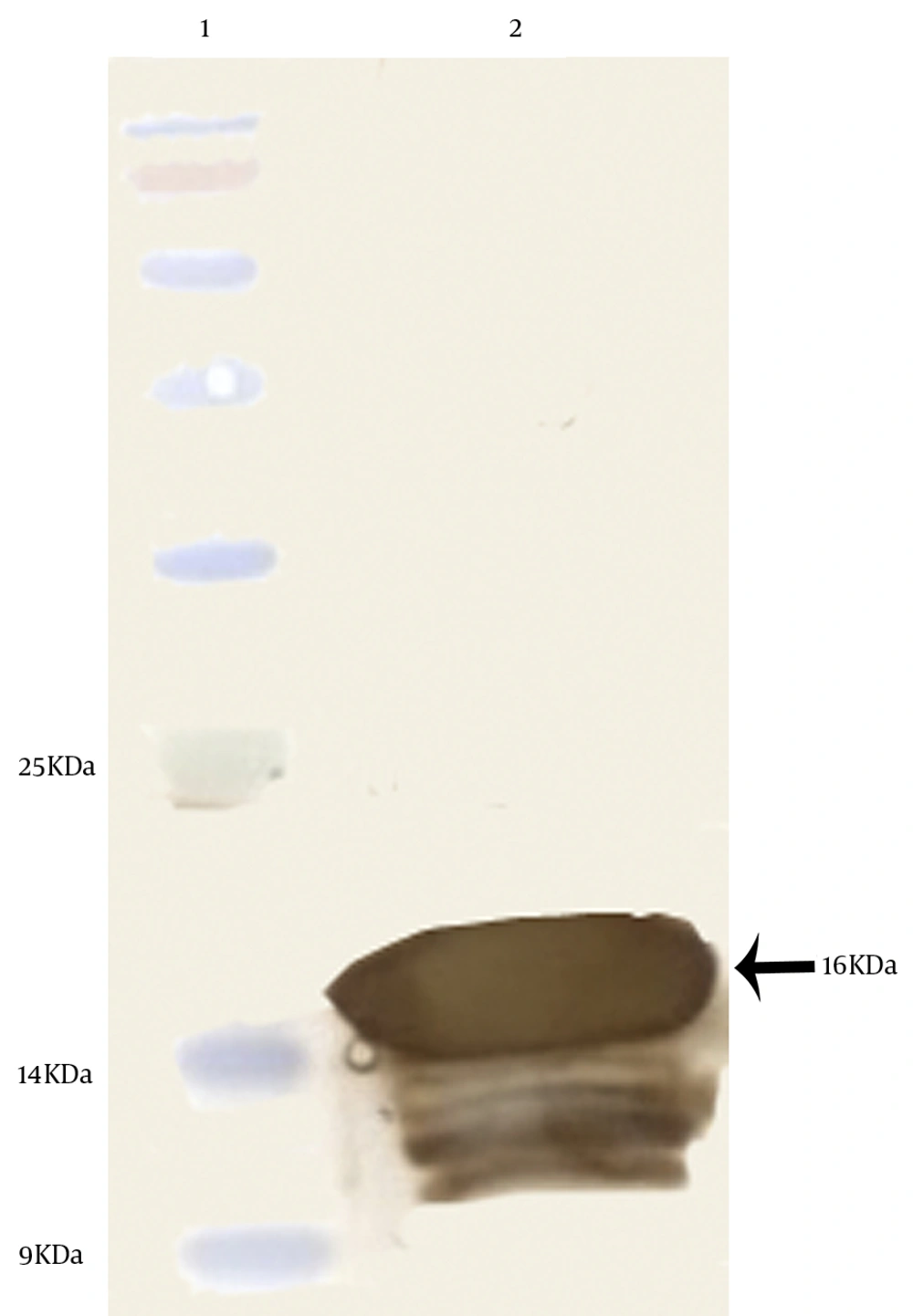
4.4. Western Blot Analysis
The western blot analysis was performed to detect the antigenicity of the expressed protein. The 16 kDa protein band, observed in the SDS-PAGE, was confirmed as PD by the western blot analysis using peroxidase-conjugated rabbit anti-mouse immunoglobulins (Figure 7).
5. Discussion
NTHi has established itself as an important human pathogen in the past several years. Effective vaccines against the Hib strains do not protect children against infections caused by the NTHi strains (4). To develop a vaccine that protects against Hib and NTHi infections, several surface-exposed H. influenzae proteins have been intensely studied. Thus, the identification of immunogenic and evaluation of the immunogenicity of immunogen protein is helpful to produce effective vaccines. PD is a potential candidate for vaccine research inasmuch as a study showed that the sequences were > 97% identical on both the nucleotide and deduced amino acid levels (13). PD (also known as LPD) is a conserved outer-membrane-associated lipoprotein in H. influenzae (18). Previous studies have shown a PD-deficient mutant, constructed by linker insertion and deletion mutagenesis, to be approximately 100-fold less virulent than its PD-expressing parental NTHi strain in inducing otitis media in an experimental rat otitis model (19). St Geme and Falkow showed that the NTHi strain lacking PD expression was less potent than the wild-type strain to cause otitis media in an experimental rat otitis model (20).
In the present study, a truncated form of PD was designed and a three-dimensional (3D)-structure protein was generated by using I-TASSER. The 3D-structure prediction for this protein and its similarity was measured by Z-scores. The analysis of the protein structure showed that the 3D structure was similar to the native protein because the Z-score was in the range of native conformations. We selected the pBAD expression system because the pBAD expression vector has a signal sequence expressing proteins in periplasm (21). The recombinant truncated PD from H. influenzae was cloned and expressed under the control of an Arabinose promoter. The obtained results showed that the pBAD expression system was very efficient. Finally, after the optimization of expression, the yields of the expression of the recombinant proteins were 4 mg per liter of bacterial cultures.
In conclusion, this project, for the first time, describes the construction of a genetically constructed truncated PD. Hence, further immunological studies are required to evaluate this protein as a novel and safe vaccine candidate against H. influenzae.